Human Genome Variation ( IF 1.0 ) Pub Date : 2019-10-18 , DOI: 10.1038/s41439-019-0079-1 Takao Hoshina , Toshiyuki Seto , Taro Shimono , Hiroaki Sakamoto , Torayuki Okuyama , Takashi Hamazaki , Toshiyuki Yamamoto
|
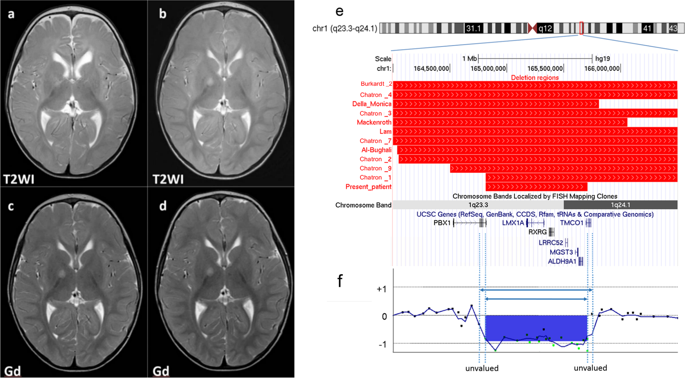
Interstitial deletions of 1q23.3q24.1 are rare. Here, chromosomal microarray testing identified a de novo microdeletion of arr[GRCh37]1q23.3q24.1(164816055_165696996) × 1 in a patient with moderate developmental delay, hearing loss, cryptorchidism, and other distinctive features. The clinical features were common to those previously reported in patients with overlapping deletions. The patient’s deletion size was 881 kb—the smallest yet reported. This therefore narrowed down the deletion responsible for the common clinical features. The deleted region included seven genes; deletion of LMX1A, RXRG, and ALDH9A1 may have caused our patient’s neurodevelopmental delay.
中文翻译:

通过识别最小的缺失来缩小负责1q23.3q24.1微缺失的区域
1q23.3q24.1的间隙删除很少见。在这里,染色体微阵列测试确定了患有中度发育延迟,听力减退,隐睾症和其他明显特征的患者的arr [GRCh37] 1q23.3q24.1(164816055_165696996)×1的从头微缺失。临床特征与先前报道的重叠缺失患者相同。患者的缺失大小为881 kb,是迄今报道的最小的缺失大小。因此,这缩小了负责共同临床特征的删除范围。缺失的区域包括七个基因。删除LMX1A,RXRG和ALDH9A1可能导致我们的病人的神经发育延迟。











































 京公网安备 11010802027423号
京公网安备 11010802027423号