Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Cell deformability heterogeneity recognition by unsupervised machine learning from in-flow motion parameters
Lab on a Chip ( IF 6.1 ) Pub Date : 2022-11-07 , DOI: 10.1039/d2lc00902a Maria Isabella Maremonti 1 , David Dannhauser 1 , Valeria Panzetta 1 , Paolo Antonio Netti 1 , Filippo Causa 1
Lab on a Chip ( IF 6.1 ) Pub Date : 2022-11-07 , DOI: 10.1039/d2lc00902a Maria Isabella Maremonti 1 , David Dannhauser 1 , Valeria Panzetta 1 , Paolo Antonio Netti 1 , Filippo Causa 1
Affiliation
|
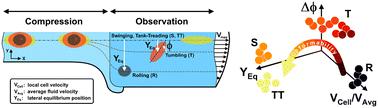
Cell deformability is a well-established marker of cell states for diagnostic purposes. However, the measurement of a wide range of different deformability levels is still challenging, especially in cancer, where a large heterogeneity of rheological/mechanical properties is present. Therefore, a simple, versatile and cost-effective recognition method for variable rheological/mechanical properties of cells is needed. Here, we introduce a new set of in-flow motion parameters capable of identifying heterogeneity among cell deformability, properly modified by the administration of drugs for cytoskeleton destabilization. Firstly, we measured cell deformability by identification of in-flow motions, rolling (R), tumbling (T), swinging (S) and tank-treading (TT), distinctively associated with cell rheological/mechanical properties. Secondly, from a pool of motion and structural cell parameters, an unsupervised machine learning approach based on principal component analysis (PCA) revealed dominant features: the local cell velocity (VCell/VAvg), the equilibrium position (YEq) and the orientation angle variation (Δφ). These motion parameters clearly defined cell clusters in terms of motion regimes corresponding to specific deformability. Such correlation is verified in a wide range of rheological/mechanical properties from the elastic cells moving like R until the almost viscous cells moving as TT. Thus, our approach shows how simple motion parameters allow cell deformability heterogeneity recognition, directly measuring rheological/mechanical properties.
中文翻译:

基于流入运动参数的无监督机器学习的细胞变形异质性识别
细胞变形能力是用于诊断目的的细胞状态的公认标志。然而,测量各种不同的可变形性水平仍然具有挑战性,尤其是在癌症中,流变学/机械特性存在很大的异质性。因此,需要一种简单、通用且具有成本效益的识别方法来识别细胞的可变流变学/机械特性。在这里,我们引入了一组新的流入运动参数,能够识别细胞变形能力之间的异质性,并通过给予细胞骨架不稳定药物进行适当修改。首先,我们通过识别流入运动、滚动 (R)、翻滚 (T)、摆动 (S) 和坦克踩踏 (TT) 来测量细胞变形能力,这些运动与细胞流变学/机械特性明显相关。第二,V Cell / V Avg),平衡位置(Y Eq)和取向角变化(Δ φ)。这些运动参数根据与特定变形能力相对应的运动机制清楚地定义了细胞簇。这种相关性在广泛的流变/机械特性中得到验证,从像 R 一样移动的弹性细胞到像 TT 一样移动的几乎粘性细胞。因此,我们的方法展示了简单的运动参数如何允许细胞变形异质性识别,直接测量流变/机械特性。
更新日期:2022-11-07
中文翻译:

基于流入运动参数的无监督机器学习的细胞变形异质性识别
细胞变形能力是用于诊断目的的细胞状态的公认标志。然而,测量各种不同的可变形性水平仍然具有挑战性,尤其是在癌症中,流变学/机械特性存在很大的异质性。因此,需要一种简单、通用且具有成本效益的识别方法来识别细胞的可变流变学/机械特性。在这里,我们引入了一组新的流入运动参数,能够识别细胞变形能力之间的异质性,并通过给予细胞骨架不稳定药物进行适当修改。首先,我们通过识别流入运动、滚动 (R)、翻滚 (T)、摆动 (S) 和坦克踩踏 (TT) 来测量细胞变形能力,这些运动与细胞流变学/机械特性明显相关。第二,V Cell / V Avg),平衡位置(Y Eq)和取向角变化(Δ φ)。这些运动参数根据与特定变形能力相对应的运动机制清楚地定义了细胞簇。这种相关性在广泛的流变/机械特性中得到验证,从像 R 一样移动的弹性细胞到像 TT 一样移动的几乎粘性细胞。因此,我们的方法展示了简单的运动参数如何允许细胞变形异质性识别,直接测量流变/机械特性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号