当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Proteomic Analysis of Azacitidine-Induced Degradation Profiles Identifies Multiple Chromatin and Epigenetic Regulators Including Uhrf1 and Dnmt1 as Sensitive to Azacitidine.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2019-02-07 , DOI: 10.1021/acs.jproteome.8b00745 Emily H Bowler 1 , Joseph Bell 1 , Nullin Divecha 1 , Paul Skipp 1 , Rob M Ewing 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2019-02-07 , DOI: 10.1021/acs.jproteome.8b00745 Emily H Bowler 1 , Joseph Bell 1 , Nullin Divecha 1 , Paul Skipp 1 , Rob M Ewing 1
Affiliation
|
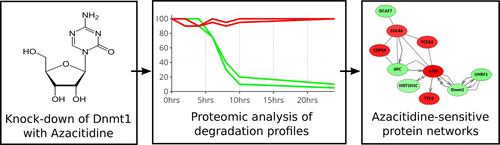
DNA methylation is a critical epigenetic modification that is established and maintained across the genome by DNA methyltransferase enzymes (Dnmts). Altered patterns of DNA methylation are a frequent occurrence in many tumor genomes, and inhibitors of Dnmts have become important epigenetic drugs. Azacitidine is a cytidine analog that is incorporated into DNA and induces the specific inhibition and proteasomal-mediated degradation of Dnmts. The downstream effects of azacitidine on CpG methylation and on gene transcription have been widely studied in many systems, but how azacitidine impacts the proteome is not well-understood. In addition, with its specific ability to induce the rapid degradation of Dnmts (in particular, the primary maintenance DNA methyltransferase, Dnmt1), it may be employed as a specific chemical knockdown for investigating the Dnmt1-associated functional or physical interactome. In this study, we use quantitative proteomics to analyze the degradation profile of proteins in the nuclear proteome of cells treated with azacitidine. We identify specific proteins as well as multiple pathways and processes that are impacted by azacitidine. The Dnmt1 interaction partner, Uhrf1, exhibits significant azacitidine-induced degradation, and this azacitidine-induced degradation is independent of the levels of Dnmt1 protein. We identify multiple other chromatin- and epigenetic-associated factors, including the bromodomain-containing transcriptional regulator, Brd2. We show that azacitidine induces highly specific perturbations of the Dnmt1-associated proteome, and while interaction partners such as Uhrf1 are sensitive to azacitidine, others such as the Dnmt1 interaction partner and stability regulator, Usp7, are not. In summary, we have conducted the first comprehensive proteomic analysis of the azacitidine-sensitive nuclear proteome, and we show how 5-azacitidine can be used as a specific probe to explore Dnmt- and chromatin-related protein networks.
中文翻译:

阿扎胞苷诱导降解曲线的蛋白质组学分析确定了多种染色质和表观遗传调节剂,包括对Uzafidine敏感的Uhrf1和Dnmt1。
DNA甲基化是一种关键的表观遗传修饰,它是通过DNA甲基转移酶(Dnmts)在整个基因组中建立和维持的。DNA甲基化模式的改变在许多肿瘤基因组中经常发生,而Dnmts抑制剂已成为重要的表观遗传药物。阿扎胞苷是胞苷类似物,其被掺入DNA中并诱导Dnmts的特异性抑制和蛋白酶体介导的降解。在许多系统中,阿扎胞苷对CpG甲基化和基因转录的下游影响已得到广泛研究,但尚未充分理解阿扎胞苷如何影响蛋白质组。此外,由于其具有诱导Dnmts(特别是初级维持DNA甲基转移酶Dnmt1)快速降解的特殊能力,它可以用作研究Dnmt1相关功能或物理相互作用组的特定化学敲低分子。在这项研究中,我们使用定量蛋白质组学来分析用阿扎胞苷处理过的细胞的核蛋白质组中蛋白质的降解情况。我们确定了受阿扎胞苷影响的特定蛋白质以及多种途径和过程。Dnmt1相互作用的伙伴,Uhrf1,显示出显着的阿扎胞苷诱导的降解,而这种阿扎胞苷诱导的降解与Dnmt1蛋白的水平无关。我们确定了多个其他染色质和表观遗传相关的因素,包括含溴结构域的转录调节剂Brd2。我们显示,阿扎胞苷可诱导Dnmt1相关蛋白质组的高特异性扰动,尽管Uhrf1等相互作用伴侣对阿扎胞苷敏感,而Dnmt1相互作用伴侣和稳定性调节剂Usp7等却不敏感。总而言之,我们已经对氮杂胞苷敏感的核蛋白质组进行了首次综合蛋白质组学分析,并且我们展示了如何将5-氮杂胞苷用作研究Dnmt和染色质相关蛋白网络的特异性探针。
更新日期:2019-02-07
中文翻译:

阿扎胞苷诱导降解曲线的蛋白质组学分析确定了多种染色质和表观遗传调节剂,包括对Uzafidine敏感的Uhrf1和Dnmt1。
DNA甲基化是一种关键的表观遗传修饰,它是通过DNA甲基转移酶(Dnmts)在整个基因组中建立和维持的。DNA甲基化模式的改变在许多肿瘤基因组中经常发生,而Dnmts抑制剂已成为重要的表观遗传药物。阿扎胞苷是胞苷类似物,其被掺入DNA中并诱导Dnmts的特异性抑制和蛋白酶体介导的降解。在许多系统中,阿扎胞苷对CpG甲基化和基因转录的下游影响已得到广泛研究,但尚未充分理解阿扎胞苷如何影响蛋白质组。此外,由于其具有诱导Dnmts(特别是初级维持DNA甲基转移酶Dnmt1)快速降解的特殊能力,它可以用作研究Dnmt1相关功能或物理相互作用组的特定化学敲低分子。在这项研究中,我们使用定量蛋白质组学来分析用阿扎胞苷处理过的细胞的核蛋白质组中蛋白质的降解情况。我们确定了受阿扎胞苷影响的特定蛋白质以及多种途径和过程。Dnmt1相互作用的伙伴,Uhrf1,显示出显着的阿扎胞苷诱导的降解,而这种阿扎胞苷诱导的降解与Dnmt1蛋白的水平无关。我们确定了多个其他染色质和表观遗传相关的因素,包括含溴结构域的转录调节剂Brd2。我们显示,阿扎胞苷可诱导Dnmt1相关蛋白质组的高特异性扰动,尽管Uhrf1等相互作用伴侣对阿扎胞苷敏感,而Dnmt1相互作用伴侣和稳定性调节剂Usp7等却不敏感。总而言之,我们已经对氮杂胞苷敏感的核蛋白质组进行了首次综合蛋白质组学分析,并且我们展示了如何将5-氮杂胞苷用作研究Dnmt和染色质相关蛋白网络的特异性探针。











































 京公网安备 11010802027423号
京公网安备 11010802027423号