npj Breast Cancer ( IF 6.5 ) Pub Date : 2019-01-18 , DOI: 10.1038/s41523-018-0101-7 Fresia Pareja 1 , Felipe C Geyer 1 , David N Brown 1 , Ana P Martins Sebastião 1 , Rodrigo Gularte-Mérida 1 , Anqi Li 1 , Marcia Edelweiss 1 , Arnaud Da Cruz Paula 1 , Pier Selenica 1 , Hannah Y Wen 1 , Achim A Jungbluth 1 , Zsuzsanna Varga 2 , Juan Palazzo 3 , Brian P Rubin 4 , Ian O Ellis 5 , Edi Brogi 1 , Emad A Rakha 5 , Britta Weigelt 1 , Jorge S Reis-Filho 1
|
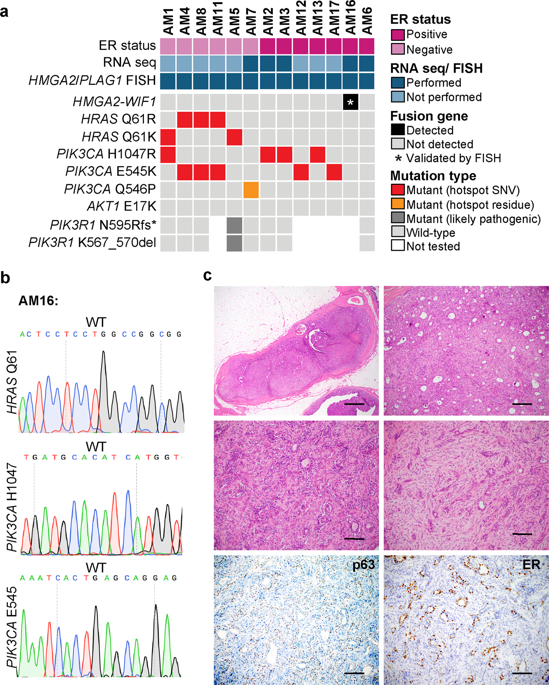
Breast adenomyoepitheliomas (AMEs) are rare epithelial-myoepithelial neoplasms that may occasionally produce myxochondroid matrix, akin to pleomorphic adenomas (PAs). Regardless of their anatomic location, PAs often harbor rearrangements involving HMGA2 or PLAG1. We have recently shown that the repertoire of somatic genetic alterations of AMEs varies according to their estrogen receptor (ER) status; whilst the majority of ER-positive AMEs display mutually exclusive PIK3CA or AKT1 hotspot mutations, up to 60% of ER-negative AMEs harbor concurrent HRAS Q61 hotspot mutations and mutations affecting either PIK3CA or PIK3R1. Here, we hypothesized that a subset of AMEs lacking these somatic genetic alterations could be underpinned by oncogenic fusion genes, in particular those involving HMGA2 or PLAG1. Therefore, we subjected 13 AMEs to RNA-sequencing for fusion discovery (n = 5) and/or fluorescence in situ hybridization (FISH) analysis for HMGA2 and PLAG1 rearrangements (n = 13). RNA-sequencing revealed an HMGA2-WIF1 fusion gene in an ER-positive AME lacking HRAS, PIK3CA and AKT1 somatic mutations. This fusion gene, which has been previously described in salivary gland PAs, results in a chimeric transcript composed of exons 1–5 of HMGA2 and exons 3–10 of WIF1. No additional in-frame fusion genes or HMGA2 or PLAG1 rearrangements were identified in the remaining AMEs analyzed. Our results demonstrate that a subset of AMEs lacking mutations affecting HRAS and PI3K pathway-related genes may harbor HMGA2-WIF1 fusion genes, suggesting that a subset of breast AMEs may be genetically related to PAs or that a subset of AMEs may originate in the context of a PA.
中文翻译:

乳腺腺肌上皮瘤中 HMGA2 和 PLAG1 重排的评估
乳腺腺肌上皮瘤(AME)是罕见的上皮肌上皮肿瘤,偶尔可能产生粘软骨样基质,类似于多形性腺瘤(PA)。无论其解剖位置如何,PA 经常包含涉及HMGA2或PLAG1的重排。我们最近发现,AME 的体细胞遗传改变根据其雌激素受体 (ER) 状态而变化。虽然大多数 ER 阳性 AME 表现出相互排斥的PIK3CA或AKT1热点突变,但高达 60% 的 ER 阴性 AME 同时存在HRAS Q61 热点突变和影响PIK3CA或PIK3R1 的突变。在这里,我们假设缺乏这些体细胞遗传改变的 AME 子集可能受到致癌融合基因的支持,特别是那些涉及HMGA2或PLAG1的基因。因此,我们对 13 个 AME 进行了 RNA 测序以进行融合发现 ( n = 5) 和/或对HMGA2和PLAG1重排 进行荧光原位杂交 (FISH) 分析( n = 13)。RNA 测序揭示了ER 阳性 AME 中存在HMGA2 - WIF1融合基因,缺乏HRAS、PIK3CA和AKT1体细胞突变。该融合基因先前已在唾液腺 PA 中描述过,会产生由HMGA2的外显子 1-5 和WIF1的外显子 3-10组成的嵌合转录本。在分析的其余 AME 中没有发现额外的框内融合基因或HMGA2或PLAG1重排。我们的结果表明,缺乏影响HRAS和 PI3K 通路相关基因突变的 AME 子集可能含有HMGA2 - WIF1融合基因,这表明乳腺 AME 子集可能与 PA 存在遗传相关性,或者 AME 子集可能起源于背景一个PA。











































 京公网安备 11010802027423号
京公网安备 11010802027423号