PLOS ONE ( IF 2.9 ) Pub Date : 2019-01-17 , DOI: 10.1371/journal.pone.0210910 Bobak D. Kechavarzi , Huanmei Wu , Thompson N. Doman
|
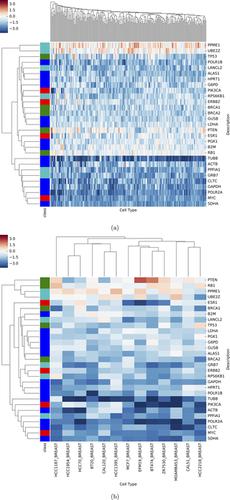
The massive genomic data from The Cancer Genome Atlas (TCGA), including proteomics data from Clinical Proteomic Tumor Analysis Consortium (CPTAC), provides a unique opportunity to study cancer systematically. While most observations are made from a single type of genomics data, we apply big data analytics and systems biology approaches by simultaneously analyzing DNA amplification, mRNA and protein abundance. Using multiple genomic profiles, we have discovered widespread dosage compensation for the extensive aneuploidy observed in TCGA breast cancer samples. We do identify 11 genes that show strong correlation across all features (DNA/mRNA/protein) analogous to that of the well-known oncogene HER2 (ERBB2). These genes are generally less well-characterized regarding their role in cancer and we advocate their further study. We also discover that shRNA knockdown of these genes has an impact on cancer cell growth, suggesting a vulnerability that could be used for cancer therapy. Our study shows the advantages of systematic big data methodologies and also provides future research directions.
中文翻译:

自下而上的综合组学分析可从TCGA中识别出广泛的剂量敏感性基因
来自癌症基因组图谱(TCGA)的大量基因组数据,包括来自临床蛋白质组学肿瘤分析协会(CPTAC)的蛋白质组学数据,为系统地研究癌症提供了独特的机会。虽然大多数观察是从单一类型的基因组数据中获得的,但我们通过同时分析DNA扩增,mRNA和蛋白质丰度,应用大数据分析和系统生物学方法。使用多个基因组图谱,我们发现了在TCGA乳腺癌样品中观察到的广泛非整倍性的广泛剂量补偿。我们确实鉴定出11个基因,这些基因在所有特征(DNA / mRNA /蛋白质)中都表现出强相关性,类似于众所周知的癌基因HER2(ERBB2)。这些基因在癌症中的作用通常不太明确,我们提倡对其进行进一步研究。我们还发现这些基因的shRNA敲低对癌细胞的生长有影响,这表明可以用于癌症治疗的脆弱性。我们的研究显示了系统的大数据方法的优势,并提供了未来的研究方向。











































 京公网安备 11010802027423号
京公网安备 11010802027423号