当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
pmartR: Quality Control and Statistics for Mass Spectrometry-Based Biological Data.
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2019-01-28 , DOI: 10.1021/acs.jproteome.8b00760 Kelly G Stratton 1 , Bobbie-Jo M Webb-Robertson 1 , Lee Ann McCue 2 , Bryan Stanfill 1 , Daniel Claborne 1 , Iobani Godinez 1 , Thomas Johansen 3 , Allison M Thompson 2 , Kristin E Burnum-Johnson 2 , Katrina M Waters 2 , Lisa M Bramer 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2019-01-28 , DOI: 10.1021/acs.jproteome.8b00760 Kelly G Stratton 1 , Bobbie-Jo M Webb-Robertson 1 , Lee Ann McCue 2 , Bryan Stanfill 1 , Daniel Claborne 1 , Iobani Godinez 1 , Thomas Johansen 3 , Allison M Thompson 2 , Kristin E Burnum-Johnson 2 , Katrina M Waters 2 , Lisa M Bramer 1
Affiliation
|
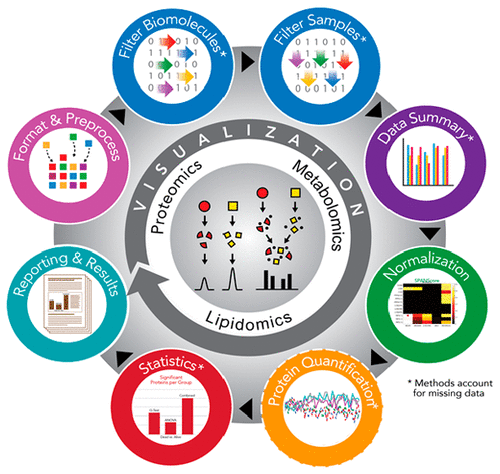
Prior to statistical analysis of mass spectrometry (MS) data, quality control (QC) of the identified biomolecule peak intensities is imperative for reducing process-based sources of variation and extreme biological outliers. Without this step, statistical results can be biased. Additionally, liquid chromatography-MS proteomics data present inherent challenges due to large amounts of missing data that require special consideration during statistical analysis. While a number of R packages exist to address these challenges individually, there is no single R package that addresses all of them. We present pmartR, an open-source R package, for QC (filtering and normalization), exploratory data analysis (EDA), visualization, and statistical analysis robust to missing data. Example analysis using proteomics data from a mouse study comparing smoke exposure to control demonstrates the core functionality of the package and highlights the capabilities for handling missing data. In particular, using a combined quantitative and qualitative statistical test, 19 proteins whose statistical significance would have been missed by a quantitative test alone were identified. The pmartR package provides a single software tool for QC, EDA, and statistical comparisons of MS data that is robust to missing data and includes numerous visualization capabilities.
中文翻译:

pmartR:基于质谱的生物数据的质量控制和统计。
在对质谱 (MS) 数据进行统计分析之前,对已识别的生物分子峰强度进行质量控制 (QC) 对于减少基于过程的变异来源和极端生物异常值至关重要。如果没有这一步,统计结果可能会有偏差。此外,由于大量缺失数据,在统计分析过程中需要特别考虑,液相色谱-MS 蛋白质组学数据也面临着固有的挑战。虽然有许多 R 包可以单独解决这些挑战,但没有一个 R 包可以解决所有这些挑战。我们推出 pmartR,一个开源 R 包,用于 QC(过滤和标准化)、探索性数据分析 (EDA)、可视化和对缺失数据稳健的统计分析。使用小鼠研究中的蛋白质组学数据进行示例分析,比较烟雾暴露与对照,展示了该软件包的核心功能,并强调了处理缺失数据的能力。特别是,通过结合定量和定性统计测试,鉴定出了 19 种蛋白质,仅靠定量测试可能会忽略这些蛋白质的统计显着性。 pmartR 软件包提供了一个用于 QC、EDA 和 MS 数据统计比较的单一软件工具,该工具对缺失数据具有鲁棒性,并包含众多可视化功能。
更新日期:2019-02-07
中文翻译:

pmartR:基于质谱的生物数据的质量控制和统计。
在对质谱 (MS) 数据进行统计分析之前,对已识别的生物分子峰强度进行质量控制 (QC) 对于减少基于过程的变异来源和极端生物异常值至关重要。如果没有这一步,统计结果可能会有偏差。此外,由于大量缺失数据,在统计分析过程中需要特别考虑,液相色谱-MS 蛋白质组学数据也面临着固有的挑战。虽然有许多 R 包可以单独解决这些挑战,但没有一个 R 包可以解决所有这些挑战。我们推出 pmartR,一个开源 R 包,用于 QC(过滤和标准化)、探索性数据分析 (EDA)、可视化和对缺失数据稳健的统计分析。使用小鼠研究中的蛋白质组学数据进行示例分析,比较烟雾暴露与对照,展示了该软件包的核心功能,并强调了处理缺失数据的能力。特别是,通过结合定量和定性统计测试,鉴定出了 19 种蛋白质,仅靠定量测试可能会忽略这些蛋白质的统计显着性。 pmartR 软件包提供了一个用于 QC、EDA 和 MS 数据统计比较的单一软件工具,该工具对缺失数据具有鲁棒性,并包含众多可视化功能。











































 京公网安备 11010802027423号
京公网安备 11010802027423号