Signal Transduction and Targeted Therapy ( IF 40.8 ) Pub Date : 2019-01-11 , DOI: 10.1038/s41392-018-0034-5 Eliseos J. Mucaki , Jonathan Z. L. Zhao , Daniel J. Lizotte , Peter K. Rogan
|
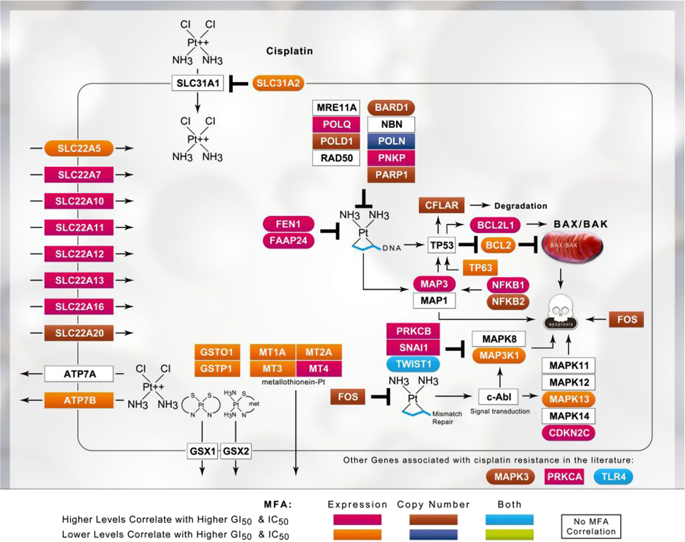
The selection of effective genes that accurately predict chemotherapy responses might improve cancer outcomes. We compare optimized gene signatures for cisplatin, carboplatin, and oxaliplatin responses in the same cell lines and validate each signature using data from patients with cancer. Supervised support vector machine learning is used to derive gene sets whose expression is related to the cell line GI50 values by backwards feature selection with cross-validation. Specific genes and functional pathways distinguishing sensitive from resistant cell lines are identified by contrasting signatures obtained at extreme and median GI50 thresholds. Ensembles of gene signatures at different thresholds are combined to reduce the dependence on specific GI50 values for predicting drug responses. The most accurate gene signatures for each platin are: cisplatin: BARD1, BCL2, BCL2L1, CDKN2C, FAAP24, FEN1, MAP3K1, MAPK13, MAPK3, NFKB1, NFKB2, SLC22A5, SLC31A2, TLR4, and TWIST1; carboplatin: AKT1, EIF3K, ERCC1, GNGT1, GSR, MTHFR, NEDD4L, NLRP1, NRAS, RAF1, SGK1, TIGD1, TP53, VEGFB, and VEGFC; and oxaliplatin: BRAF, FCGR2A, IGF1, MSH2, NAGK, NFE2L2, NQO1, PANK3, SLC47A1, SLCO1B1, and UGT1A1. Data from The Cancer Genome Atlas (TCGA) patients with bladder, ovarian, and colorectal cancer were used to test the cisplatin, carboplatin, and oxaliplatin signatures, resulting in 71.0%, 60.2%, and 54.5% accuracies in predicting disease recurrence and 59%, 61%, and 72% accuracies in predicting remission, respectively. One cisplatin signature predicted 100% of recurrence in non-smoking patients with bladder cancer (57% disease-free; N = 19), and 79% recurrence in smokers (62% disease-free; N = 35). This approach should be adaptable to other studies of chemotherapy responses, regardless of the drug or cancer types.
中文翻译:

借助生物化学启发式机器学习预测对铂类化疗药物的反应
选择能准确预测化疗反应的有效基因可能会改善癌症的预后。我们比较了同一细胞系中顺铂,卡铂和奥沙利铂反应的优化基因签名,并使用来自癌症患者的数据验证了每个签名。监督支持向量机学习用于通过交叉验证的反向特征选择来得出其表达与细胞系GI 50值相关的基因集。通过对比在极端和中等GI 50阈值下获得的特征来鉴定区分敏感细胞与耐药细胞系的特定基因和功能途径。将不同阈值的基因签名组合在一起以减少对特定GI 50的依赖性预测药物反应的值。每个铂的最准确的基因标记是:顺铂:BARD1,BCL2,BCL2L1,CDKN2C,FAAP24,FEN1,MAP3K1,MAPK13,MAPK3,NFKB1,NFKB2,SLC22A5,SLC31A2,TLR4和TWIST1 ; 卡铂:AKT1,EIF3K,ERCC1,GNGT1,GSR,MTHFR,NEDD4L,NLRP1,NRAS,RAF1,SGK1,TIGD1,TP53,VEGFB和VEGFC; 和奥沙利铂:BRAF,FCGR2A,IGF1,MSH2,NAGK,NFE2L2,NQO1,PANK3,SLC47A1,SLCO1B1和UGT1A1。来自患有膀胱癌,卵巢癌和结肠直肠癌的癌症基因组图谱(TCGA)患者的数据用于测试顺铂,卡铂和奥沙利铂的特征,在预测疾病复发和预测疾病复发方面的准确度分别为71.0%,60.2%和54.5%,而59% ,分别在预测缓解方面的准确度为61%和72%。一种顺铂信号预测非吸烟膀胱癌患者的复发率为100%(无疾病; 57%;N = 19);吸烟者复发率为79%(无疾病; 62%;N = 35)。不论药物或癌症类型如何,该方法均应适用于其他化学疗法的研究。











































 京公网安备 11010802027423号
京公网安备 11010802027423号