当前位置:
X-MOL 学术
›
Genet. Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Detection of structural variation using target captured next-generation sequencing data for genetic diagnostic testing.
Genetics in Medicine ( IF 6.6 ) Pub Date : 2018-12-19 , DOI: 10.1038/s41436-018-0397-6 Wenbo Mu 1 , Bing Li 1 , Sitao Wu 1 , Jefferey Chen 1 , Divya Sain 1 , Dong Xu 1 , Mary Helen Black 1 , Rachid Karam 1 , Katrina Gillespie 1 , Kelly D Farwell Hagman 1 , Lucia Guidugli 1 , Melissa Pronold 1 , Aaron Elliott 1 , Hsiao-Mei Lu 1
Genetics in Medicine ( IF 6.6 ) Pub Date : 2018-12-19 , DOI: 10.1038/s41436-018-0397-6 Wenbo Mu 1 , Bing Li 1 , Sitao Wu 1 , Jefferey Chen 1 , Divya Sain 1 , Dong Xu 1 , Mary Helen Black 1 , Rachid Karam 1 , Katrina Gillespie 1 , Kelly D Farwell Hagman 1 , Lucia Guidugli 1 , Melissa Pronold 1 , Aaron Elliott 1 , Hsiao-Mei Lu 1
Affiliation
|
PURPOSE
Structural variation (SV) is associated with inherited diseases. Next-generation sequencing (NGS) is an efficient method for SV detection because of its high-throughput, low cost, and base-pair resolution. However, due to lack of standard NGS protocols and a limited number of clinical samples with pathogenic SVs, comprehensive standards for SV detection, interpretation, and reporting are to be established.
METHODS
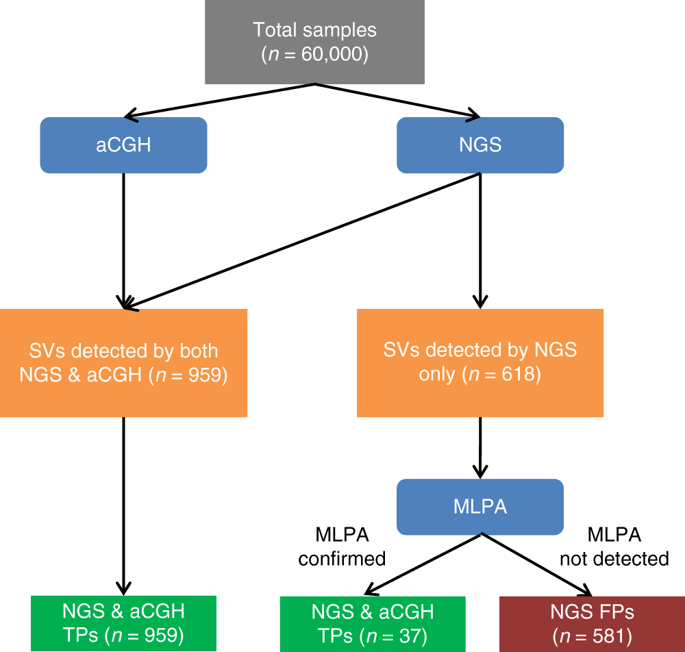
We performed SV assessment on 60,000 clinical samples tested with hereditary cancer NGS panels spanning 48 genes. To evaluate NGS results, NGS and orthogonal methods were used separately in a blinded fashion for SV detection in all samples.
RESULTS
A total of 1,037 SVs in coding sequence (CDS) or untranslated regions (UTRs) and 30,847 SVs in introns were detected and validated. Across all variant types, NGS shows 100% sensitivity and 99.9% specificity. Overall, 64% of CDS/UTR SVs were classified as pathogenic/likely pathogenic, and five deletions/duplications were reclassified as pathogenic using breakpoint information from NGS.
CONCLUSION
The SVs presented here can be used as a valuable resource for clinical research and diagnostics. The data illustrate NGS as a powerful tool for SV detection. Application of NGS and confirmation technologies in genetic testing ensures delivering accurate and reliable results for diagnosis and patient care.
中文翻译:

使用目标捕获的下一代测序数据检测结构变异以进行基因诊断测试。
目的 结构变异 (SV) 与遗传性疾病有关。新一代测序 (NGS) 因其高通量、低成本和碱基对分辨率而成为 SV 检测的有效方法。然而,由于缺乏标准的 NGS 方案和具有致病性 SV 的临床样本数量有限,需要建立 SV 检测、解释和报告的综合标准。方法 我们对 60,000 个临床样本进行了 SV 评估,这些样本使用了跨越 48 个基因的遗传性癌症 NGS 面板进行测试。为了评估 NGS 结果,NGS 和正交方法以盲法分别用于所有样品中的 SV 检测。结果 共检测和验证了编码序列 (CDS) 或非翻译区 (UTR) 中的 1,037 个 SV 和内含子中的 30,847 个 SV。在所有变体类型中,NGS 显示 100% 的敏感性和 99.9% 的特异性。总体而言,64% 的 CDS/UTR SV 被分类为致病/可能致病,并且使用来自 NGS 的断点信息将五个缺失/重复重新分类为致病。结论 这里介绍的 SV 可以用作临床研究和诊断的宝贵资源。这些数据说明 NGS 是一种强大的 SV 检测工具。在基因检测中应用 NGS 和确认技术可确保为诊断和患者护理提供准确可靠的结果。结论 这里介绍的 SV 可以用作临床研究和诊断的宝贵资源。这些数据说明 NGS 是一种强大的 SV 检测工具。在基因检测中应用 NGS 和确认技术可确保为诊断和患者护理提供准确可靠的结果。结论 这里介绍的 SV 可以用作临床研究和诊断的宝贵资源。这些数据说明 NGS 是一种强大的 SV 检测工具。在基因检测中应用 NGS 和确认技术可确保为诊断和患者护理提供准确可靠的结果。
更新日期:2019-01-26
中文翻译:

使用目标捕获的下一代测序数据检测结构变异以进行基因诊断测试。
目的 结构变异 (SV) 与遗传性疾病有关。新一代测序 (NGS) 因其高通量、低成本和碱基对分辨率而成为 SV 检测的有效方法。然而,由于缺乏标准的 NGS 方案和具有致病性 SV 的临床样本数量有限,需要建立 SV 检测、解释和报告的综合标准。方法 我们对 60,000 个临床样本进行了 SV 评估,这些样本使用了跨越 48 个基因的遗传性癌症 NGS 面板进行测试。为了评估 NGS 结果,NGS 和正交方法以盲法分别用于所有样品中的 SV 检测。结果 共检测和验证了编码序列 (CDS) 或非翻译区 (UTR) 中的 1,037 个 SV 和内含子中的 30,847 个 SV。在所有变体类型中,NGS 显示 100% 的敏感性和 99.9% 的特异性。总体而言,64% 的 CDS/UTR SV 被分类为致病/可能致病,并且使用来自 NGS 的断点信息将五个缺失/重复重新分类为致病。结论 这里介绍的 SV 可以用作临床研究和诊断的宝贵资源。这些数据说明 NGS 是一种强大的 SV 检测工具。在基因检测中应用 NGS 和确认技术可确保为诊断和患者护理提供准确可靠的结果。结论 这里介绍的 SV 可以用作临床研究和诊断的宝贵资源。这些数据说明 NGS 是一种强大的 SV 检测工具。在基因检测中应用 NGS 和确认技术可确保为诊断和患者护理提供准确可靠的结果。结论 这里介绍的 SV 可以用作临床研究和诊断的宝贵资源。这些数据说明 NGS 是一种强大的 SV 检测工具。在基因检测中应用 NGS 和确认技术可确保为诊断和患者护理提供准确可靠的结果。











































 京公网安备 11010802027423号
京公网安备 11010802027423号