npj Genomic Medicine ( IF 4.7 ) Pub Date : 2018-12-18 , DOI: 10.1038/s41525-018-0072-5 Hindrek Teder , Mariann Koel , Priit Paluoja , Tatjana Jatsenko , Kadri Rekker , Triin Laisk-Podar , Viktorija Kukuškina , Agne Velthut-Meikas , Olga Fjodorova , Maire Peters , Juha Kere , Andres Salumets , Priit Palta , Kaarel Krjutškov
|
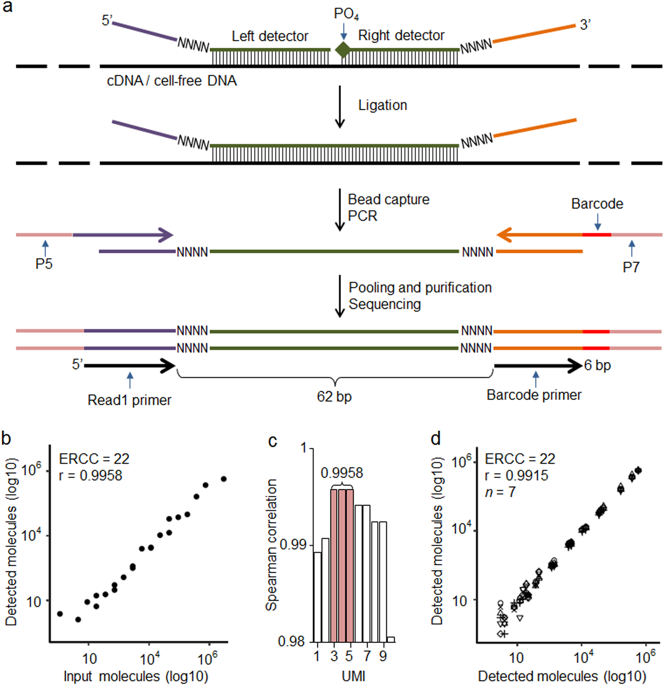
Targeted next-generation sequencing (NGS) methods have become essential in medical research and diagnostics. In addition to NGS sensitivity and high-throughput capacity, precise biomolecule counting based on unique molecular identifier (UMI) has potential to increase biomolecule detection accuracy. Although UMIs are widely used in basic research its introduction to clinical assays is still in progress. Here, we present a robust and cost-effective TAC-seq (Targeted Allele Counting by sequencing) method that uses UMIs to estimate the original molecule counts of mRNAs, microRNAs, and cell-free DNA. We applied TAC-seq in three different clinical applications and compared the results with standard NGS. RNA samples extracted from human endometrial biopsies were analyzed using previously described 57 mRNA-based receptivity biomarkers and 49 selected microRNAs at different expression levels. Cell-free DNA aneuploidy testing was based on cell line (47,XX, +21) genomic DNA. TAC-seq mRNA profiling showed identical clustering results to transcriptome RNA sequencing, and microRNA detection demonstrated significant reduction in amplification bias, allowing to determine minor expression changes between different samples that remained undetermined by standard NGS. The mimicking experiment for cell-free DNA fetal aneuploidy analysis showed that TAC-seq can be applied to count highly fragmented DNA, detecting significant (p = 7.6 × 10−4) excess of chromosome 21 molecules at 10% fetal fraction level. Based on three proof-of-principle applications we demonstrate that TAC-seq is an accurate and highly potential biomarker profiling method for advanced medical research and diagnostics.
中文翻译:

TAC-seq:靶向DNA和RNA测序,可进行精确的生物标志物分子计数
有针对性的下一代测序(NGS)方法在医学研究和诊断中已变得至关重要。除了NGS灵敏度和高通量能力外,基于独特分子识别符(UMI)的精确生物分子计数还具有提高生物分子检测精度的潜力。尽管UMI在基础研究中被广泛使用,但其对临床测定的介绍仍在进行中。在这里,我们介绍了一种强大且具有成本效益的TAC-seq(测序定向等位基因计数)方法,该方法使用UMI来估计mRNA,microRNA和无细胞DNA的原始分子数。我们将TAC-seq应用于三种不同的临床应用中,并将结果与标准NGS进行了比较。使用先前描述的57种基于mRNA的接受性生物标记物和49种选择的不同表达水平的microRNA,分析了从人子宫内膜活检组织中提取的RNA样品。无细胞DNA非整倍性测试基于细胞系(47,XX,+21)基因组DNA。TAC-seq mRNA分析显示出与转录组RNA测序相同的聚类结果,microRNA检测证明扩增偏差显着降低,从而可以确定标准NGS仍未确定的不同样品之间的微小表达变化。无细胞DNA胎儿非整倍性分析的模拟实验表明,TAC-seq可用于计数高度片段化的DNA,检测出显着的 TAC-seq mRNA分析显示出与转录组RNA测序相同的聚类结果,microRNA检测证明扩增偏差显着降低,从而可以确定标准NGS仍未确定的不同样品之间的微小表达变化。无细胞DNA胎儿非整倍性分析的模拟实验表明,TAC-seq可用于计数高度片段化的DNA,检测出显着的 TAC-seq mRNA分析显示出与转录组RNA测序相同的聚类结果,microRNA检测证明扩增偏差显着降低,从而可以确定标准NGS仍未确定的不同样品之间的微小表达变化。无细胞DNA胎儿非整倍性分析的模拟实验表明,TAC-seq可用于计数高度片段化的DNA,检测出显着的p = 7.6×10 -4)以10%的胎儿分数水平过量的21号染色体分子。基于三项原理验证应用,我们证明TAC-seq是一种用于高级医学研究和诊断的准确且潜力巨大的生物标志物分析方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号