当前位置:
X-MOL 学术
›
Integr. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Protein structure networks provide insight into active site flexibility in esterase/lipases from the carnivorous plant Drosera capensis.
Integrative Biology ( IF 1.5 ) Pub Date : 2018-12-19 , DOI: 10.1039/c8ib00140e Vy T Duong 1 , Megha H Unhelkar , John E Kelly , Suhn H Kim , Carter T Butts , Rachel W Martin
Integrative Biology ( IF 1.5 ) Pub Date : 2018-12-19 , DOI: 10.1039/c8ib00140e Vy T Duong 1 , Megha H Unhelkar , John E Kelly , Suhn H Kim , Carter T Butts , Rachel W Martin
Affiliation
|
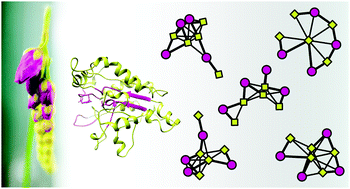
In plants, esterase/lipases perform transesterification reactions, playing an important role in the synthesis of useful molecules, such as those comprising the waxy coatings of leaf surfaces. Plant genomes and transcriptomes have provided a wealth of data about expression patterns and the circumstances under which these enzymes are upregulated, e.g. pathogen defense and response to drought; however, predicting their functional characteristics from genomic or transcriptome data is challenging due to weak sequence conservation among the diverse members of this group. Although functional sequence blocks mediating enzyme activity have been identified, progress to date has been hampered by the paucity of information on the structural relationships among these regions and how they affect substrate specificity. Here we present methodology for predicting overall protein flexibility and active site flexibility based on molecular modeling and analysis of protein structure networks (PSNs). We define two new types of specialized PSNs: sequence region networks (SRNs) and active site networks (ASNs), which provide parsimonious representations of molecular structure in reference to known features of interest. Our approach, intended as an aid to target selection for poorly characterized enzyme classes, is demonstrated for 26 previously uncharacterized esterase/lipases from the genome of the carnivorous plant Drosera capensis and validated using a case/control design. Analysis of the network relationships among functional blocks and among the chemical moieties making up the catalytic triad reveals potentially functionally significant differences that are not apparent from sequence analysis alone.
中文翻译:

蛋白质结构网络提供了对肉食植物茅膏菜酯酶/脂肪酶活性位点灵活性的深入了解。
在植物中,酯酶/脂肪酶进行酯交换反应,在有用分子的合成中发挥重要作用,例如构成叶子表面蜡质涂层的分子。植物基因组和转录组提供了有关表达模式和这些酶上调的环境的大量数据,例如病原体防御和对干旱的反应;然而,由于该组不同成员之间的序列保守性较弱,从基因组或转录组数据预测其功能特征具有挑战性。尽管已经鉴定了介导酶活性的功能序列块,但由于缺乏关于这些区域之间的结构关系以及它们如何影响底物特异性的信息,迄今为止的进展受到阻碍。在这里,我们提出了基于分子建模和蛋白质结构网络(PSN)分析来预测整体蛋白质灵活性和活性位点灵活性的方法。我们定义了两种新型的专用 PSN:序列区域网络(SRN)和活性位点网络(ASN),它们根据已知的感兴趣特征提供分子结构的简洁表示。我们的方法旨在帮助目标选择特征较差的酶类,针对来自食肉植物 Drosera capensis 基因组的 26 种以前未表征的酯酶/脂肪酶进行了论证,并使用病例/对照设计进行了验证。对功能块之间以及构成催化三联体的化学部分之间的网络关系的分析揭示了潜在的功能上显着的差异,这些差异仅通过序列分析是不明显的。
更新日期:2018-12-05
中文翻译:

蛋白质结构网络提供了对肉食植物茅膏菜酯酶/脂肪酶活性位点灵活性的深入了解。
在植物中,酯酶/脂肪酶进行酯交换反应,在有用分子的合成中发挥重要作用,例如构成叶子表面蜡质涂层的分子。植物基因组和转录组提供了有关表达模式和这些酶上调的环境的大量数据,例如病原体防御和对干旱的反应;然而,由于该组不同成员之间的序列保守性较弱,从基因组或转录组数据预测其功能特征具有挑战性。尽管已经鉴定了介导酶活性的功能序列块,但由于缺乏关于这些区域之间的结构关系以及它们如何影响底物特异性的信息,迄今为止的进展受到阻碍。在这里,我们提出了基于分子建模和蛋白质结构网络(PSN)分析来预测整体蛋白质灵活性和活性位点灵活性的方法。我们定义了两种新型的专用 PSN:序列区域网络(SRN)和活性位点网络(ASN),它们根据已知的感兴趣特征提供分子结构的简洁表示。我们的方法旨在帮助目标选择特征较差的酶类,针对来自食肉植物 Drosera capensis 基因组的 26 种以前未表征的酯酶/脂肪酶进行了论证,并使用病例/对照设计进行了验证。对功能块之间以及构成催化三联体的化学部分之间的网络关系的分析揭示了潜在的功能上显着的差异,这些差异仅通过序列分析是不明显的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号