当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Ketoreductase Domain Dysfunction Expands Chemodiversity: Malyngamide Biosynthesis in the Cyanobacterium Okeania hirsuta.
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2018-12-03 , DOI: 10.1021/acschembio.8b00910 Nathan A Moss 1 , Tiago Leão 1 , Michael R Rankin 2 , Tyler M McCullough 2 , Pingping Qu 3 , Anton Korobeynikov 4 , Janet L Smith 2 , Lena Gerwick 1 , William H Gerwick 1, 5
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2018-12-03 , DOI: 10.1021/acschembio.8b00910 Nathan A Moss 1 , Tiago Leão 1 , Michael R Rankin 2 , Tyler M McCullough 2 , Pingping Qu 3 , Anton Korobeynikov 4 , Janet L Smith 2 , Lena Gerwick 1 , William H Gerwick 1, 5
Affiliation
|
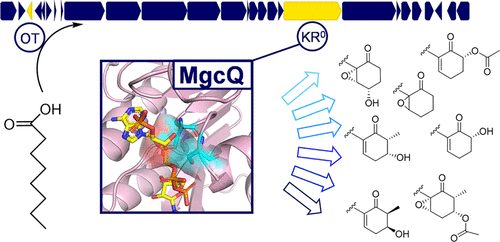
Dozens of type A malyngamides, principally identified by a decorated six-membered cyclohexanone headgroup and methoxylated lyngbic acid tail, have been isolated over several decades. Their environmental sources include macro- and microbiotic organisms, including sea hares, red alga, and cyanobacterial assemblages, but the true producing organism has remained enigmatic. Many type A analogues display potent bioactivity in human-health related assays, spurring an interest in this molecular class and its biosynthetic pathway. Here, we present the discovery of the type A malyngamide biosynthetic pathway in the first sequenced genome of the cyanobacterial genus Okeania. Bioinformatic analysis of two cultured Okeania genome assemblies identified 62 and 68 kb polyketide synthase/nonribosomal peptide synthetase (PKS/NRPS) pathways with unusual loading and termination genes. NMR data of malyngamide C acetate derived from 13C-substrate-fed cultures provided evidence that an intact octanoate moiety is transferred to the first KS module via a LipM homologue originally associated with lipoic acid metabolism and implicated an inactive ketoreductase (KR0) as critical for six-membered ring formation, a hallmark of the malyngamide family. Phylogenetic analysis and homology modeling of the penultimate KR0 domain inferred structural cofactor binding and active site alterations as contributory to domain dysfunction, which was confirmed by recombinant protein expression and NADPH binding assay. The carbonyl retained from this KR0 ultimately enables an intramolecular Knoevenagel condensation to form the characteristic cyclohexanone ring. Understanding this critical step allows assignment of a biosynthetic model for all type A malyngamides, whereby well-characterized tailoring modifications explain the surprising proliferation and diversity of analogues.
中文翻译:

酮还原酶结构域功能异常扩展了化学多样性:蓝藻Okeania hirsuta中的麦芽酰胺生物合成。
几十年来,已分离出数十种主要由修饰的六元环己酮头基和甲氧基化的次要酸尾巴鉴定的数十种A型麦芽酰胺。它们的环境来源包括大型和微生物生物,包括海兔,红藻和蓝细菌群落,但真正的产生生物仍是谜。许多A型类似物在与人类健康相关的测定中显示出强大的生物活性,引起了对该分子类别及其生物合成途径的兴趣。在这里,我们介绍了蓝藻属Okeania的第一个测序基因组中A型麦芽酰胺生物合成途径的发现。对两个培养的Okeania基因组装配体的生物信息学分析确定了62和68 kb的聚酮化合物合酶/非核糖体肽合成酶(PKS / NRPS)途径具有异常的加载和终止基因。来源于13C底物喂养的培养物中的乙酸丙二酰胺C的NMR数据提供了一个证据,即一个完整的辛酸酯部分通过最初与硫辛酸代谢相关的LipM同源物转移到了第一个KS模块,并暗示了非活性的酮还原酶(KR0)对于六个关键元环形成,是麦芽酰胺家族的标志。倒数第二个KR0结构域的系统进化分析和同源性建模推断结构辅因子结合和活性位点改变是结构域功能障碍的原因,这已通过重组蛋白表达和NADPH结合测定得以证实。从该KR 0保留的羰基最终使分子内的Knoevenagel缩合形成特征性的环己酮环。了解了这一关键步骤后,就可以为所有A型麦芽酰胺分配一个生物合成模型,从而充分表征定制修饰可以解释类似物的令人惊讶的增殖和多样性。
更新日期:2018-11-16
中文翻译:

酮还原酶结构域功能异常扩展了化学多样性:蓝藻Okeania hirsuta中的麦芽酰胺生物合成。
几十年来,已分离出数十种主要由修饰的六元环己酮头基和甲氧基化的次要酸尾巴鉴定的数十种A型麦芽酰胺。它们的环境来源包括大型和微生物生物,包括海兔,红藻和蓝细菌群落,但真正的产生生物仍是谜。许多A型类似物在与人类健康相关的测定中显示出强大的生物活性,引起了对该分子类别及其生物合成途径的兴趣。在这里,我们介绍了蓝藻属Okeania的第一个测序基因组中A型麦芽酰胺生物合成途径的发现。对两个培养的Okeania基因组装配体的生物信息学分析确定了62和68 kb的聚酮化合物合酶/非核糖体肽合成酶(PKS / NRPS)途径具有异常的加载和终止基因。来源于13C底物喂养的培养物中的乙酸丙二酰胺C的NMR数据提供了一个证据,即一个完整的辛酸酯部分通过最初与硫辛酸代谢相关的LipM同源物转移到了第一个KS模块,并暗示了非活性的酮还原酶(KR0)对于六个关键元环形成,是麦芽酰胺家族的标志。倒数第二个KR0结构域的系统进化分析和同源性建模推断结构辅因子结合和活性位点改变是结构域功能障碍的原因,这已通过重组蛋白表达和NADPH结合测定得以证实。从该KR 0保留的羰基最终使分子内的Knoevenagel缩合形成特征性的环己酮环。了解了这一关键步骤后,就可以为所有A型麦芽酰胺分配一个生物合成模型,从而充分表征定制修饰可以解释类似物的令人惊讶的增殖和多样性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号