当前位置:
X-MOL 学术
›
Mol. Omics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Combined multivariate analysis and machine learning reveals a predictive module of metabolic stress response in Arabidopsis thaliana
Molecular Omics ( IF 3.0 ) Pub Date : 2018-11-02 , DOI: 10.1039/c8mo00095f Lisa Fürtauer 1 , Alice Pschenitschnigg , Helene Scharkosi , Wolfram Weckwerth , Thomas Nägele
Molecular Omics ( IF 3.0 ) Pub Date : 2018-11-02 , DOI: 10.1039/c8mo00095f Lisa Fürtauer 1 , Alice Pschenitschnigg , Helene Scharkosi , Wolfram Weckwerth , Thomas Nägele
Affiliation
|
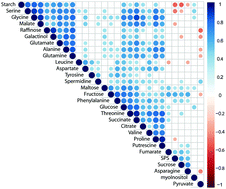
Abiotic stress exposure of plants induces metabolic reprogramming which is tightly regulated by signalling cascades connecting transcriptional with translational and metabolic regulation. Complexity of such interconnected metabolic networks impedes the functional understanding of molecular plant stress response compromising the design of breeding strategies and biotechnological processes. Thus, defining a molecular network to enable the prediction of a plant's stress mode will improve the understanding of stress responsive biochemical regulation and will yield novel molecular targets for technological application. Arabidopsis wild type plants and two mutant lines with deficiency in sucrose or starch metabolism were grown under ambient and combined cold/high light stress conditions. Stress-induced dynamics of the primary metabolome and the proteome were quantified by mass spectrometry. Wild type data were used to train a machine learning algorithm to classify mutant lines under control and stress conditions. Multivariate analysis and classification identified a module consisting of 23 proteins enabling the reliable prediction of combined temperature/high light stress conditions. 18 of these 23 proteins displayed putative protein–protein interactions connecting transcriptional regulation with regulation of primary and secondary metabolism. The identified stress-responsive core module supports prediction of complex biochemical regulation under changing environmental conditions.
中文翻译:

多变量分析和机器学习相结合揭示了拟南芥代谢应激反应的预测模块
植物的非生物胁迫暴露会诱导代谢重编程,而代谢重编程受到连接转录与翻译和代谢调节的信号级联的严格调节。这种相互关联的代谢网络的复杂性阻碍了对分子植物胁迫反应的功能理解,从而损害了育种策略和生物技术过程的设计。因此,定义一个分子网络来预测植物的胁迫模式将提高对胁迫响应生化调节的理解,并将产生用于技术应用的新分子目标。拟南芥野生型植物和两个蔗糖或淀粉代谢缺陷的突变株系在环境和冷/强光联合胁迫条件下生长。通过质谱法对应激诱导的初级代谢组和蛋白质组的动态进行量化。野生型数据用于训练机器学习算法,以在控制和应激条件下对突变株系进行分类。多变量分析和分类确定了一个由 23 种蛋白质组成的模块,能够可靠地预测组合温度/强光应激条件。这 23 种蛋白质中的 18 种显示出假定的蛋白质-蛋白质相互作用,将转录调节与初级和次级代谢的调节联系起来。确定的应激响应核心模块支持在不断变化的环境条件下预测复杂的生化调节。
更新日期:2018-12-03
中文翻译:

多变量分析和机器学习相结合揭示了拟南芥代谢应激反应的预测模块
植物的非生物胁迫暴露会诱导代谢重编程,而代谢重编程受到连接转录与翻译和代谢调节的信号级联的严格调节。这种相互关联的代谢网络的复杂性阻碍了对分子植物胁迫反应的功能理解,从而损害了育种策略和生物技术过程的设计。因此,定义一个分子网络来预测植物的胁迫模式将提高对胁迫响应生化调节的理解,并将产生用于技术应用的新分子目标。拟南芥野生型植物和两个蔗糖或淀粉代谢缺陷的突变株系在环境和冷/强光联合胁迫条件下生长。通过质谱法对应激诱导的初级代谢组和蛋白质组的动态进行量化。野生型数据用于训练机器学习算法,以在控制和应激条件下对突变株系进行分类。多变量分析和分类确定了一个由 23 种蛋白质组成的模块,能够可靠地预测组合温度/强光应激条件。这 23 种蛋白质中的 18 种显示出假定的蛋白质-蛋白质相互作用,将转录调节与初级和次级代谢的调节联系起来。确定的应激响应核心模块支持在不断变化的环境条件下预测复杂的生化调节。










































 京公网安备 11010802027423号
京公网安备 11010802027423号