Fungal Biology Reviews ( IF 5.7 ) Pub Date : 2018-10-19 , DOI: 10.1016/j.fbr.2018.09.001 Lene Lange , Bo Pilgaard , Florian-Alexander Herbst , Peter Kamp Busk , Frank Gleason , Anders Gorm Pedersen
|
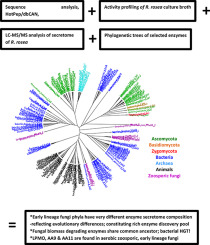
The aim of this study was to elucidate the evolution of enzyme secretome of early lineage fungi to contribute to resolving the basal part of Fungal Kingdom and pave the way for industrial evaluation of their unique enzymes. By combining results of advanced sequence analysis with secretome mass spectrometry and phylogenetic trees, we provide evidence for that plant cell wall degrading enzymes of higher fungi share a common ancestor with enzymes from aerobic ancient fungi. Sequence analysis (HotPep, confirmed by dbCAN-HMM models) enabled prediction of enzyme function directly from sequence. For the first time, oxidative enzymes are described here in early lineage fungi (Chytridiomycota & Cryptomycota), which supports the conceptually new understanding that fungal LPMOs were also present in the early evolution of the Fungal Kingdom. Phylogenetic analysis of fungal AA9 proteins suggests an LPMO-common-ancestor with Ascomycetes and Basidiomycetes and describes a new clade of AA9s. We identified two very strong biomass degraders, Rhizophlyctis rosea (soil-inhabiting) and Neocallimastix californiae (rumen), with a rich spectrum of cellulolytic, xylanolytic and pectinolytic enzymes, characteristically including several different enzymes with the same function. Their secretome composition suggests horizontal gene transfer was involved in transition to terrestrial and rumen habitats. Methods developed for recombinant production and protein characterization of enzymes from zoosporic fungi pave the way for biotechnological exploitation of unique enzymes from early lineage fungi with potential to contribute to improved biomass conversion. The phyla of ancient fungi through evolution have developed to be very different and together they constitute a rich enzyme discovery pool.
中文翻译:

真菌生物质降解酶的起源:早期谱系真菌的酶的进化,多样性和功能
这项研究的目的是阐明早期谱系真菌的酶分泌组的进化,以有助于解决真菌王国的基础部分,并为对其独特酶的工业评价铺平道路。通过将高级序列分析的结果与促分泌素质谱和系统进化树相结合,我们提供了证据,表明高级真菌的植物细胞壁降解酶与有氧古代真菌的酶具有共同的祖先。序列分析(HotPep,由dbCAN-HMM模型确认)可以直接从序列中预测酶的功能。首次在早期谱系真菌(Chytridiomycota和Cryptomycota)中描述了氧化酶,这在概念上支持了新的认识,即真菌LPMOs也存在于真菌王国的早期进化中。真菌AA9蛋白的系统发生分析表明,LPMO与子囊菌和担子菌共生,并描述了AA9s的新进化枝。我们确定了两个非常强大的生物质降解剂,玫瑰根霉(土壤寄居)和加利福尼亚新callimastix(瘤胃),具有丰富的纤维素分解酶,木聚糖分解酶和果胶分解酶谱,特征在于包括几种具有相同功能的不同酶。他们的分泌组组成表明,水平基因转移参与了向陆地和瘤胃生境的过渡。为游动孢子真菌的酶的重组生产和蛋白质表征而开发的方法,为从早期谱系真菌中独特酶的生物技术开发铺平了道路,这可能有助于改善生物量转化。通过进化,古代真菌的门已经发展非常不同,它们共同构成了丰富的酶发现库。









































 京公网安备 11010802027423号
京公网安备 11010802027423号