当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Modeling gene-regulatory networks to describe cell fate transitions and predict master regulators
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2018-08-02 , DOI: 10.1038/s41540-018-0066-z Pierre-Etienne Cholley , Julien Moehlin , Alexia Rohmer , Vincent Zilliox , Samuel Nicaise , Hinrich Gronemeyer , Marco Antonio Mendoza-Parra
中文翻译:

对基因调控网络进行建模以描述细胞命运的转变并预测主调控因子
更新日期:2019-05-16
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2018-08-02 , DOI: 10.1038/s41540-018-0066-z Pierre-Etienne Cholley , Julien Moehlin , Alexia Rohmer , Vincent Zilliox , Samuel Nicaise , Hinrich Gronemeyer , Marco Antonio Mendoza-Parra
|
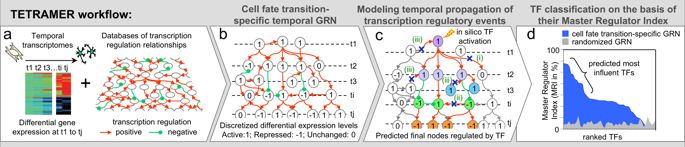
Complex organisms originate from and are maintained by the information encoded in the genome. A major challenge of systems biology is to develop algorithms that describe the dynamic regulation of genome functions from large omics datasets. Here, we describe TETRAMER, which reconstructs gene-regulatory networks from temporal transcriptome data during cell fate transitions to predict “master” regulators by simulating cascades of temporal transcription-regulatory events.
中文翻译:

对基因调控网络进行建模以描述细胞命运的转变并预测主调控因子
复杂生物起源于基因组中编码的信息并由其维护。系统生物学的一个主要挑战是开发描述来自大型组学数据集的基因组功能的动态调节的算法。在这里,我们描述了TETRAMER,它可以通过模拟时间转录调控事件的级联,从细胞命运转变过程中的时间转录组数据重建基因调控网络,从而预测“主要”调控因子。











































 京公网安备 11010802027423号
京公网安备 11010802027423号