npj Genomic Medicine ( IF 4.7 ) Pub Date : 2018-07-09 , DOI: 10.1038/s41525-018-0053-8 Michelle M. Clark , Zornitza Stark , Lauge Farnaes , Tiong Y. Tan , Susan M. White , David Dimmock , Stephen F. Kingsmore
|
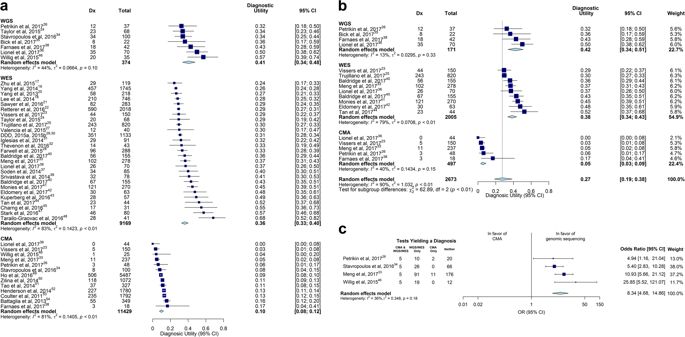
Genetic diseases are leading causes of childhood mortality. Whole-genome sequencing (WGS) and whole-exome sequencing (WES) are relatively new methods for diagnosing genetic diseases, whereas chromosomal microarray (CMA) is well established. Here we compared the diagnostic utility (rate of causative, pathogenic, or likely pathogenic genotypes in known disease genes) and clinical utility (proportion in whom medical or surgical management was changed by diagnosis) of WGS, WES, and CMA in children with suspected genetic diseases by systematic review of the literature (January 2011–August 2017) and meta-analysis, following MOOSE/PRISMA guidelines. In 37 studies, comprising 20,068 children, diagnostic utility of WGS (0.41, 95% CI 0.34–0.48, I2 = 44%) and WES (0.36, 95% CI 0.33–0.40, I2 = 83%) were qualitatively greater than CMA (0.10, 95% CI 0.08–0.12, I2 = 81%). Among studies published in 2017, the diagnostic utility of WGS was significantly greater than CMA (P < 0.0001, I2 = 13% and I2 = 40%, respectively). Among studies featuring within-cohort comparisons, the diagnostic utility of WES was significantly greater than CMA (P < 0.001, I2 = 36%). The diagnostic utility of WGS and WES were not significantly different. In studies featuring within-cohort comparisons of WGS/WES, the likelihood of diagnosis was significantly greater for trios than singletons (odds ratio 2.04, 95% CI 1.62–2.56, I2 = 12%; P < 0.0001). Diagnostic utility of WGS/WES with hospital-based interpretation (0.42, 95% CI 0.38–0.45, I2 = 48%) was qualitatively higher than that of reference laboratories (0.29, 95% CI 0.27–0.31, I2 = 49%); this difference was significant among studies published in 2017 (P < .0001, I2 = 22% and I2 = 26%, respectively). The clinical utility of WGS (0.27, 95% CI 0.17–0.40, I2 = 54%) and WES (0.17, 95% CI 0.12–0.24, I2 = 76%) were higher than CMA (0.06, 95% CI 0.05–0.07, I2 = 42%); this difference was significant for WGS vs CMA (P < 0.0001). In conclusion, in children with suspected genetic diseases, the diagnostic and clinical utility of WGS/WES were greater than CMA. Subgroups with higher WGS/WES diagnostic utility were trios and those receiving hospital-based interpretation. WGS/WES should be considered a first-line genomic test for children with suspected genetic diseases.
中文翻译:

疑似遗传病儿童基因组,外显子组测序和染色体微阵列诊断和临床应用的荟萃分析
遗传疾病是导致儿童死亡的主要原因。全基因组测序(WGS)和全外显子测序(WES)是诊断遗传疾病的相对较新的方法,而染色体微阵列(CMA)则已建立起来。在这里,我们比较了WGS,WES和CMA在可疑遗传儿童中的诊断效用(已知疾病基因中的致病性,致病性或可能致病性基因型的发生率)和临床效用(通过诊断改变医疗或外科治疗的比例)通过按照MOOSE / PRISMA指南对文献(2011年1月至2017年8月)进行系统回顾和进行荟萃分析来对这些疾病进行研究。在37个研究中,包括20,068名儿童,WGS(0.41,95%CI 0.34–0.48,I 2 = 44%)和WES(0.36,95%CI 0.33–0.40,I2 = 83%)在质量上大于CMA(0.10,95%CI 0.08-0.12,I 2 = 81%)。在2017年发表的研究中,WGS的诊断效用显着高于CMA(P <0.0001,I 2 = 13%和I 2 = 40%)。在以组内比较为特征的研究中,WES的诊断效用显着高于CMA(P <0.001,I 2 = 36%)。WGS和WES的诊断效用没有显着差异。在以WGS / WES进行队列内比较的研究中,三重症的诊断可能性明显高于单身者(优势比2.04、95%CI 1.62–2.56,I 2 = 12%;P <0.0001)。WGS / WES具有医院解释的诊断效用(0.42,95%CI 0.38–0.45,I 2 = 48%)在质量上高于参考实验室(0.29,95%CI 0.27–0.31,I 2 = 49% ); 在2017年发表的研究中,这一差异是显着的(P <.0001,I 2 = 22%和I 2 = 26%)。WGS(0.27,95%CI 0.17–0.40,I 2 = 54%)和WES(0.17,95%CI 0.12–0.24,I 2 = 76%)的临床效用均高于CMA(0.06,95 %CI 0.05) –0.07,我2 = 42%);对于WGS与CMA,这一差异是显着的(P <0.0001)。总之,在怀疑患有遗传病的儿童中,WGS / WES的诊断和临床效用大于CMA。具有较高WGS / WES诊断效用的亚组是三重奏,而那些接受基于医院的解释。WGS / WES应该被认为是可疑遗传疾病儿童的一线基因组测试。











































 京公网安备 11010802027423号
京公网安备 11010802027423号