npj Regenerative Medicine ( IF 6.4 ) Pub Date : 2018-05-29 , DOI: 10.1038/s41536-018-0049-0 Benjamin L King 1, 2, 3 , Michael C Rosenstein 1, 4 , Ashley M Smith 1 , Christina A Dykeman 1 , Grace A Smith 2, 5 , Viravuth P Yin 1, 3
|
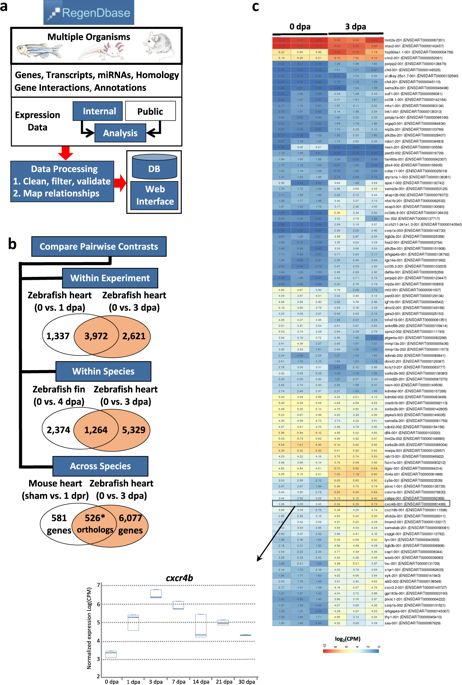
Regeneration is an endogenous process of tissue repair that culminates in complete restoration of tissue and organ function. While regenerative capacity in mammals is limited to select tissues, lower vertebrates like zebrafish and salamanders are endowed with the capacity to regenerate entire limbs and most adult tissues, including heart muscle. Numerous profiling studies have been conducted using these research models in an effort to identify the genetic circuits that accompany tissue regeneration. Most of these studies, however, are confined to an individual injury model and/or research organism and focused primarily on protein encoding transcripts. Here we describe RegenDbase, a new database with the functionality to compare and contrast gene regulatory pathways within and across tissues and research models. RegenDbase combines pipelines that integrate analysis of noncoding RNAs in combination with protein encoding transcripts. We created RegenDbase with a newly generated comprehensive dataset for adult zebrafish heart regeneration combined with existing microarray and RNA-sequencing studies on multiple injured tissues. In this current release, we detail microRNA–mRNA regulatory circuits and the biological processes these interactions control during the early stages of heart regeneration. Moreover, we identify known and putative novel lncRNAs and identify their potential target genes based on proximity searches. We postulate that these candidate factors underscore robust regenerative capacity in lower vertebrates. RegenDbase provides a systems-level analysis of tissue regeneration genetic circuits across injury and animal models and addresses the growing need to understand how noncoding RNAs influence these changes in gene expression.
中文翻译:

RegenDbase:跨多个类群的组织再生回路的非编码 RNA 调节的比较数据库
再生是组织修复的内源性过程,最终导致组织和器官功能的完全恢复。虽然哺乳动物的再生能力仅限于选定的组织,但斑马鱼和蝾螈等低等脊椎动物却具有再生整个四肢和大多数成体组织(包括心肌)的能力。人们利用这些研究模型进行了大量的分析研究,试图确定伴随组织再生的遗传回路。然而,大多数这些研究仅限于个体损伤模型和/或研究生物体,并且主要集中于蛋白质编码转录本。在这里,我们描述了 RegenDbase,这是一个新数据库,具有比较和对比组织和研究模型内部和之间的基因调控途径的功能。RegenDbase 结合了将非编码 RNA 分析与蛋白质编码转录本结合起来的管道。我们使用新生成的成年斑马鱼心脏再生综合数据集,结合现有的针对多种损伤组织的微阵列和 RNA 测序研究,创建了 RegenDbase。在当前版本中,我们详细介绍了 microRNA-mRNA 调节回路以及这些相互作用在心脏再生早期阶段控制的生物过程。此外,我们还识别已知和推定的新型 lncRNA,并基于邻近搜索识别其潜在靶基因。我们假设这些候选因素强调了低等脊椎动物强大的再生能力。RegenDbase 提供了跨损伤和动物模型的组织再生遗传回路的系统级分析,并满足了了解非编码 RNA 如何影响基因表达变化的日益增长的需求。











































 京公网安备 11010802027423号
京公网安备 11010802027423号