当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Precise Probing of Residue Roles by NRPS Code Swapping: Mutation, Enzymatic Characterization, Modeling, and Substrate Promiscuity of Aryl Acid Adenylation Domains.
Biochemistry ( IF 2.9 ) Pub Date : 2020-01-15 , DOI: 10.1021/acs.biochem.9b00748 Fumihiro Ishikawa 1 , Maya Nohara 1 , Shinya Nakamura 2 , Isao Nakanishi 2 , Genzoh Tanabe 1
Biochemistry ( IF 2.9 ) Pub Date : 2020-01-15 , DOI: 10.1021/acs.biochem.9b00748 Fumihiro Ishikawa 1 , Maya Nohara 1 , Shinya Nakamura 2 , Isao Nakanishi 2 , Genzoh Tanabe 1
Affiliation
|
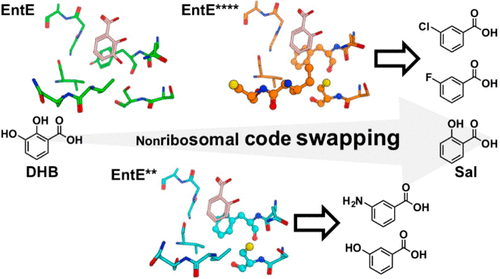
Aryl acids are most commonly found in iron-scavenging siderophores but are not limited to them. The nonribosomal peptide synthetase (NRPS) codes of aryl acids remain poorly elucidated relative to those of amino acids. Here, we defined more precisely the role of active-site residues in aryl acid adenylation domains (A-domains) by gradually grafting the NRPS codes used for salicylic acid (Sal) into an archetypal aryl acid A-domain, EntE [specific for the substrate 2,3-dihydroxybenzoic acid (DHB)]. Enzyme kinetics and modeling studies of these EntE variants demonstrated that the NRPS code residues at positions 236, 240, and 339 collectively regulate the substrate specificity toward DHB and Sal. Furthermore, the EntE variants exhibited the ability to activate the non-native aryl acids 3-hydroxybenzoic acid, 3-aminobenzoic acid, 3-fluorobenzoic acid, and 3-chlorobenzoic acid. These studies enhance our knowledge of the NRPS codes of aryl acids and could be exploited to reprogram aryl acid A-domains for non-native aryl acids.
中文翻译:

通过NRPS代码交换精确探测残基作用:芳基丙烯酸腺苷酸化域的突变,酶促表征,建模和底物混杂。
芳基酸最常见于除铁铁载体中,但不限于此。相对于氨基酸而言,芳基酸的非核糖体肽合成酶(NRPS)代码仍然难以阐明。在这里,我们通过逐步将用于水杨酸(Sal)的NRPS代码逐渐嫁接到原型芳基酸A域EntE [特异于底物2,3-二羟基苯甲酸(DHB)]。这些EntE变体的酶动力学和模型研究表明,位置236、240和339处的NRPS代码残基共同调节了对DHB和Sal的底物特异性。此外,EntE变体还具有激活非天然芳酸3-羟基苯甲酸,3-氨基苯甲酸,3-氟苯甲酸的能力,和3-氯苯甲酸。这些研究增强了我们对芳基酸的NRPS编码的认识,可用于重新编程非天然芳基酸的芳基酸A结构域。
更新日期:2020-01-15
中文翻译:

通过NRPS代码交换精确探测残基作用:芳基丙烯酸腺苷酸化域的突变,酶促表征,建模和底物混杂。
芳基酸最常见于除铁铁载体中,但不限于此。相对于氨基酸而言,芳基酸的非核糖体肽合成酶(NRPS)代码仍然难以阐明。在这里,我们通过逐步将用于水杨酸(Sal)的NRPS代码逐渐嫁接到原型芳基酸A域EntE [特异于底物2,3-二羟基苯甲酸(DHB)]。这些EntE变体的酶动力学和模型研究表明,位置236、240和339处的NRPS代码残基共同调节了对DHB和Sal的底物特异性。此外,EntE变体还具有激活非天然芳酸3-羟基苯甲酸,3-氨基苯甲酸,3-氟苯甲酸的能力,和3-氯苯甲酸。这些研究增强了我们对芳基酸的NRPS编码的认识,可用于重新编程非天然芳基酸的芳基酸A结构域。



























 京公网安备 11010802027423号
京公网安备 11010802027423号