The ISME Journal ( IF 10.8 ) Pub Date : 2020-01-02 , DOI: 10.1038/s41396-019-0577-7 Jenny M Broniewski 1 , Sean Meaden 1 , Steve Paterson 2 , Angus Buckling 1 , Edze R Westra 1
|
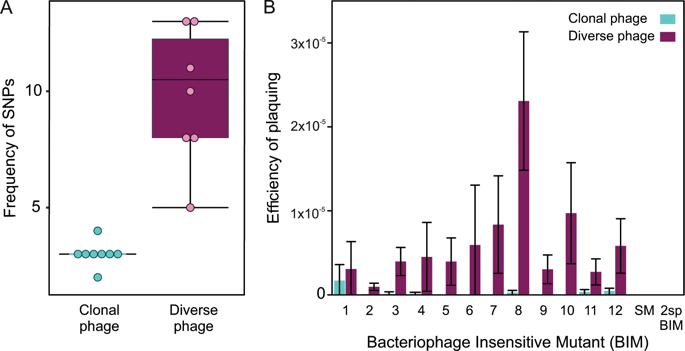
CRISPR-Cas adaptive immune systems are found in bacteria and archaea and provide defence against phage by inserting phage-derived sequences into CRISPR loci on the host genome to provide sequence specific immunological memory against re-infection. Under laboratory conditions the bacterium Pseudomonas aeruginosa readily evolves the high levels of CRISPR-based immunity against clonal populations of its phage DMS3vir, which in turn causes rapid extinction of the phage. However, in nature phage populations are likely to be more genetically diverse, which could theoretically impact the frequency at which CRISPR-based immunity evolves which in turn can alter phage persistence over time. Here we experimentally test these ideas and found that a smaller proportion of infected bacterial populations evolved CRISPR-based immunity against more genetically diverse phage populations, with the majority of the population evolving a sm preventing phage adsorption and providing generalised defence against a broader range of phage genotypes. However, those cells that do evolve CRISPR-based immunity in response to infection with more genetically diverse phage acquire greater numbers of CRISPR memory sequences in order to resist a wider range of phage genotypes. Despite differences in bacterial resistance evolution, the rates of phage extinction were similar in the context of clonal and diverse phage infections suggesting selection for CRISPR-based immunity or sm-based resistance plays a relatively minor role in the ecological dynamics in this study. Collectively, these data help to understand the drivers of CRISPR-based immunity and their consequences for bacteria-phage coexistence, and, more broadly, when generalised defences will be favoured over more specific defences.
中文翻译:

噬菌体遗传多样性对细菌抗性进化的影响。
CRISPR-Cas 适应性免疫系统存在于细菌和古生菌中,并通过将噬菌体衍生的序列插入宿主基因组上的 CRISPR 基因座中来提供针对噬菌体的防御,以提供针对再感染的序列特异性免疫记忆。在实验室条件下,细菌铜绿假单胞菌很容易进化出针对其噬菌体 DMS3vir 的克隆群体的高水平基于 CRISPR 的免疫,这反过来又导致噬菌体的快速灭绝。然而,在自然界中,噬菌体种群可能在遗传上更加多样化,这在理论上可能会影响基于 CRISPR 的免疫进化的频率,而这反过来又会随着时间的推移改变噬菌体的持久性。在这里,我们对这些想法进行了实验测试,发现一小部分受感染的细菌群体进化出基于 CRISPR 的免疫能力,以对抗基因更多样化的噬菌体群体,其中大多数群体进化出一种 sm 防止噬菌体吸附并提供针对更广泛的噬菌体的全面防御基因型。然而,那些确实进化出基于 CRISPR 的免疫以响应更多遗传多样性的噬菌体感染的细胞会获得更多数量的 CRISPR 记忆序列,以抵抗更广泛的噬菌体基因型。尽管细菌抗性进化存在差异,但在克隆和多种噬菌体感染的背景下,噬菌体的灭绝率相似,这表明选择基于 CRISPR 的免疫或基于 sm 的抗性在本研究的生态动力学中起相对较小的作用。总的来说,这些数据有助于了解基于 CRISPR 的免疫的驱动因素及其对细菌-噬菌体共存的影响,以及更广泛地说,当广义防御将优于更具体的防御时。尽管细菌抗性进化存在差异,但在克隆和多种噬菌体感染的背景下,噬菌体的灭绝率相似,这表明选择基于 CRISPR 的免疫或基于 sm 的抗性在本研究的生态动力学中起相对较小的作用。总的来说,这些数据有助于了解基于 CRISPR 的免疫的驱动因素及其对细菌-噬菌体共存的影响,以及更广泛地说,当广义防御将优于更具体的防御时。尽管细菌抗性进化存在差异,但在克隆和多种噬菌体感染的背景下,噬菌体的灭绝率相似,这表明选择基于 CRISPR 的免疫或基于 sm 的抗性在本研究的生态动力学中起相对较小的作用。总的来说,这些数据有助于了解基于 CRISPR 的免疫的驱动因素及其对细菌-噬菌体共存的影响,以及更广泛地说,当广义防御将优于更具体的防御时。











































 京公网安备 11010802027423号
京公网安备 11010802027423号