当前位置:
X-MOL 学术
›
Lung Cancer
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Clearing of circulating tumour DNA predicts clinical response to first line tyrosine kinase inhibitors in advanced epidermal growth factor receptor mutated non-small cell lung cancer.
Lung Cancer ( IF 4.5 ) Pub Date : 2019-12-30 , DOI: 10.1016/j.lungcan.2019.12.016 Eva Boysen Fynboe Ebert 1 , Tine McCulloch 2 , Karin Holmskov Hansen 3 , Hanne Linnet 4 , Boe Sorensen 5 , Peter Meldgaard 1
Lung Cancer ( IF 4.5 ) Pub Date : 2019-12-30 , DOI: 10.1016/j.lungcan.2019.12.016 Eva Boysen Fynboe Ebert 1 , Tine McCulloch 2 , Karin Holmskov Hansen 3 , Hanne Linnet 4 , Boe Sorensen 5 , Peter Meldgaard 1
Affiliation
|
OBJECTIVES
Epidermal growth factor receptor (EGFR) mutations confer sensitivity to tyrosine kinase inhibitors (TKIs) in non-small cell lung cancer (NSCLC). However, a subset of patients has limited or no response. We investigated the initial dynamics of EGFR mutations detected in circulating tumour DNA (ctDNA) during treatment as a predictive marker of outcome.
METHODS
A total of 225 patients with advanced EGFR mutated NSCLC were included for consecutive blood sampling in this prospective multicentre study. Out of these, 146 patients received first line TKI and had a baseline blood sample available for EGFR mutation testing with the Cobas® EGFR mutation test V2. For examinations on clearing and clinical outcome, 98 patients who had detectable ctDNA at baseline and at least one follow-up blood sample were included.
RESULTS
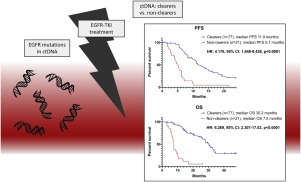
For patients with EGFR mutations present in plasma at baseline, clearing of mutations from the blood during first line TKI served as a positive predictor for objective response rate (p = 0.0008), progression-free survival (PFS) (p < 0.0001) and overall survival (OS) (p < 0.0001). This was seen both for patients who cleared the ctDNA within the first 7 weeks of treatment and patients who cleared the ctDNA at a slower pace. Baseline mutation presence was a negative predictor for PFS (p = 0.0069) and OS (p = 0.0340).
CONCLUSION
The current study is the first to confirm, in a sizeable Caucasian cohort, that clearing of EGFR mutations predict outcome to first line TKI in patients with EGFR mutated NSCLC.
中文翻译:

清除循环肿瘤DNA可以预测晚期表皮生长因子受体突变的非小细胞肺癌对一线酪氨酸激酶抑制剂的临床反应。
目的表皮生长因子受体(EGFR)突变赋予非小细胞肺癌(NSCLC)中的酪氨酸激酶抑制剂(TKIs)敏感性。但是,部分患者的反应有限或没有反应。我们调查了治疗过程中循环肿瘤DNA(ctDNA)中检测到的EGFR突变的初步动态,将其作为预后的预测指标。方法该前瞻性多中心研究共纳入225例晚期EGFR突变的NSCLC患者,以进行连续血液采样。其中,有146名患者接受了第一线TKI手术,并具有可通过EGFR突变测试V2进行EGFR突变测试的基线血液样本。为了检查清除率和临床结果,纳入了98例基线时可检测ctDNA并至少采集了一次随访血样的患者。结果对于基线时血浆中存在EGFR突变的患者,在一线TKI期间从血液中清除突变可作为客观反应率(p = 0.0008),无进展生存期(PFS)(p <0.0001)和总生存(OS)(p <0.0001)。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者的一线TKI预后。一线TKI期间从血液中清除突变可作为客观反应率(p = 0.0008),无进展生存期(PFS)(p <0.0001)和总生存期(OS)(p <0.0001)的阳性预测指标。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者的一线TKI预后。一线TKI期间从血液中清除突变可作为客观反应率(p = 0.0008),无进展生存期(PFS)(p <0.0001)和总生存期(OS)(p <0.0001)的阳性预测指标。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者的一线TKI预后。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者的一线TKI预后。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者一线TKI的预后。
更新日期:2019-12-31
中文翻译:

清除循环肿瘤DNA可以预测晚期表皮生长因子受体突变的非小细胞肺癌对一线酪氨酸激酶抑制剂的临床反应。
目的表皮生长因子受体(EGFR)突变赋予非小细胞肺癌(NSCLC)中的酪氨酸激酶抑制剂(TKIs)敏感性。但是,部分患者的反应有限或没有反应。我们调查了治疗过程中循环肿瘤DNA(ctDNA)中检测到的EGFR突变的初步动态,将其作为预后的预测指标。方法该前瞻性多中心研究共纳入225例晚期EGFR突变的NSCLC患者,以进行连续血液采样。其中,有146名患者接受了第一线TKI手术,并具有可通过EGFR突变测试V2进行EGFR突变测试的基线血液样本。为了检查清除率和临床结果,纳入了98例基线时可检测ctDNA并至少采集了一次随访血样的患者。结果对于基线时血浆中存在EGFR突变的患者,在一线TKI期间从血液中清除突变可作为客观反应率(p = 0.0008),无进展生存期(PFS)(p <0.0001)和总生存(OS)(p <0.0001)。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者的一线TKI预后。一线TKI期间从血液中清除突变可作为客观反应率(p = 0.0008),无进展生存期(PFS)(p <0.0001)和总生存期(OS)(p <0.0001)的阳性预测指标。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者的一线TKI预后。一线TKI期间从血液中清除突变可作为客观反应率(p = 0.0008),无进展生存期(PFS)(p <0.0001)和总生存期(OS)(p <0.0001)的阳性预测指标。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者的一线TKI预后。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者的一线TKI预后。对于在治疗的前7周内清除ctDNA的患者和以较慢的速度清除ctDNA的患者都可以看到这一点。基线突变的存在是PFS(p = 0.0069)和OS(p = 0.0340)的阴性预测因子。结论当前的研究是首次在相当大的白种人队列中证实,EGFR突变的清除可预测EGFR突变的NSCLC患者一线TKI的预后。











































 京公网安备 11010802027423号
京公网安备 11010802027423号