当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Sequential Multidimensional Analysis Algorithm for Aptamer Identification based on Structure Analysis and Machine Learning.
Analytical Chemistry ( IF 6.7 ) Pub Date : 2020-01-08 , DOI: 10.1021/acs.analchem.9b05203 Jia Song 1 , Yuan Zheng 1 , Mengjiao Huang 2 , Lingling Wu 1 , Wei Wang 1 , Zhi Zhu 2 , Yanling Song 1, 2 , Chaoyong Yang 1, 2
Analytical Chemistry ( IF 6.7 ) Pub Date : 2020-01-08 , DOI: 10.1021/acs.analchem.9b05203 Jia Song 1 , Yuan Zheng 1 , Mengjiao Huang 2 , Lingling Wu 1 , Wei Wang 1 , Zhi Zhu 2 , Yanling Song 1, 2 , Chaoyong Yang 1, 2
Affiliation
|
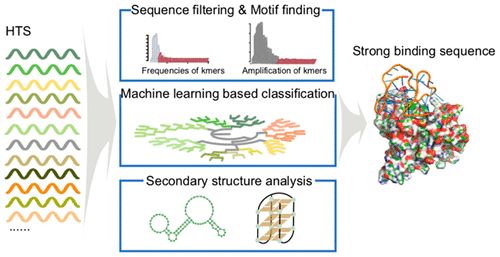
Molecular recognition ligands are of great significance in many fields, but our ability to develop new recognition molecules remains to be expanded. Here, we developed a Sequential Multidimensional Analysis algoRiThm for aptamer discovery (SMART-Aptamer) from high-throughput sequencing (HTS) data of SELEX libraries based on multilevel structure analysis and unsupervised machine learning to discover nucleic acid recognition ligands with high accuracy and efficiency. We validated SMART-Aptamer with three sets of HTS data from screening pools against hESCs, EpCAM, and CSV. High affinity aptamers for all three targets were successfully obtained, and the results revealed that SMART-Aptamer is able to pick out high affinity aptamers with low false positive and negative rates. With the advantages of accuracy, efficiency, and robustness, SMART-Aptamer represents a paradigm-shift strategy for the discovery of binding ligands for a variety of biomedical applications.
中文翻译:

基于结构分析和机器学习的序列式适体识别多维分析算法。
分子识别配体在许多领域都具有重要意义,但我们开发新的识别分子的能力仍有待扩展。在这里,我们基于多层次结构分析和无监督机器学习,基于SELEX库的高通量测序(HTS)数据,开发了用于适体发现(SMART-Aptamer)的顺序多维分析算法(SMART-Aptamer),以发现高精度和高效的核酸识别配体。我们用来自hESC,EpCAM和CSV筛选池的三组HTS数据验证了SMART-Aptamer。成功获得了针对所有三个靶标的高亲和力适体,结果表明,SMART-Aptamer能够筛选出假阳性和阴性率低的高亲和力适体。凭借准确性,效率和鲁棒性的优势,
更新日期:2020-01-09
中文翻译:

基于结构分析和机器学习的序列式适体识别多维分析算法。
分子识别配体在许多领域都具有重要意义,但我们开发新的识别分子的能力仍有待扩展。在这里,我们基于多层次结构分析和无监督机器学习,基于SELEX库的高通量测序(HTS)数据,开发了用于适体发现(SMART-Aptamer)的顺序多维分析算法(SMART-Aptamer),以发现高精度和高效的核酸识别配体。我们用来自hESC,EpCAM和CSV筛选池的三组HTS数据验证了SMART-Aptamer。成功获得了针对所有三个靶标的高亲和力适体,结果表明,SMART-Aptamer能够筛选出假阳性和阴性率低的高亲和力适体。凭借准确性,效率和鲁棒性的优势,











































 京公网安备 11010802027423号
京公网安备 11010802027423号