International Journal of Hygiene and Environmental Health ( IF 4.5 ) Pub Date : 2019-12-24 , DOI: 10.1016/j.ijheh.2019.113440 Marta Rusiñol 1 , Sandra Martínez-Puchol 1 , Natalia Timoneda 2 , Xavier Fernández-Cassi 1 , Alba Pérez-Cataluña 3 , Ana Fernández-Bravo 3 , Laura Moreno-Mesonero 4 , Yolanda Moreno 4 , Jose Luís Alonso 4 , Maria José Figueras 3 , Josep Francesc Abril 5 , Sílvia Bofill-Mas 1 , Rosina Girones 1
|
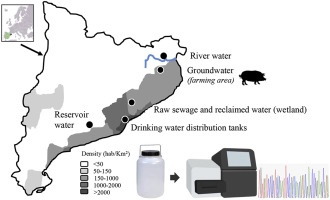
Viruses (e.g., noroviruses and hepatitis A and E virus), bacteria (e.g., Salmonella spp. and pathogenic Escherichia coli) and protozoa (e.g., Cryptosporidium parvum and Giardia intestinalis) are well-known contributors to food-borne illnesses linked to contaminated fresh produce. As agricultural irrigation increases the total amount of water used annually, reclaimed water is a good alternative to reduce dependency on conventional irrigation water sources. European guidelines have established acceptable concentrations of certain pathogens and/or indicators in irrigation water, depending on the irrigation system used and the irrigated crop. However, the incidences of food-borne infections are known to be underestimated and all the different pathogens contributing to these infections are not known. Next-generation sequencing (NGS) enables the determination of the viral, bacterial and protozoan populations present in a water sample, providing an opportunity to detect emerging pathogens and develop improved tools for monitoring the quality of irrigation water. This is a descriptive study of the virome, bacteriome and parasitome present in different irrigation water sources. We applied the same concentration method for all the studied samples and specific metagenomic approaches to characterize both DNA and RNA viruses, bacteria and protozoa.
In general, most of the known viral species corresponded to plant viruses and bacteriophages. Viral diversity in river water varied over the year, with higher bacteriophage prevalences during the autumn and winter. Reservoir water contained Enterobacter cloacae, an opportunistic human pathogen and an indicator of fecal contamination, as well as Naegleria australiensis and Naegleria clarki. Hepatitis E virus and Naegleria fowleri, emerging human pathogens, were detected in groundwater. Reclaimed water produced in a constructed wetland system presented a virome and bacteriome that resembled those of freshwater samples (river and reservoir water). Viral, bacterial and protozoan pathogens were occasionally detected in the different irrigation water sources included in this study, justifying the use of improved NGS techniques to get a comprehensive evaluation of microbial species and potential environmental health hazards associated to irrigation water.
中文翻译:

灌溉水中病毒,细菌和原生动物的元基因组分析。
病毒(例如诺如病毒以及甲型和戊型肝炎病毒),细菌(例如沙门氏菌和致病性大肠杆菌)和原生动物(例如小隐孢子虫和贾第鞭毛虫))是与被污染的新鲜农产品有关的食源性疾病的知名因素。由于农业灌溉每年增加用水总量,因此再生水是减少对常规灌溉水源依赖的好选择。欧洲指南已根据所使用的灌溉系统和灌溉作物确定了灌溉水中某些病原体和/或指标的可接受浓度。但是,已知食源性感染的发生率被低估了,导致这些感染的所有不同病原体也是未知的。下一代测序(NGS)可以确定水样中存在的病毒,细菌和原生动物种群,提供机会发现新出现的病原体,并开发改进的工具来监测灌溉水的质量。这是对不同灌溉水源中的病毒,细菌和寄生虫的描述性研究。我们对所有研究样品采用了相同的浓缩方法,并采用了特定的宏基因组学方法来表征DNA和RNA病毒,细菌和原生动物。
通常,大多数已知的病毒物种对应于植物病毒和噬菌体。一年中河水中的病毒多样性有所不同,秋季和冬季的噬菌体流行率较高。水库中的水含有阴沟肠杆菌,机会性人类病原体和粪便污染的指示剂,以及南极纳格勒菌和克拉克纳格勒菌。戊型肝炎病毒和家禽在地下水中发现了新兴的人类病原体。在人工湿地系统中产生的再生水具有类似于淡水样品(河流和水库水)的病毒和细菌组。在本研究中,在不同的灌溉水源中偶尔检测到病毒,细菌和原生动物病原体,证明使用改进的NGS技术对微生物种类和与灌溉水相关的潜在环境健康危害进行全面评估是合理的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号