Nature Machine Intelligence ( IF 18.8 ) Pub Date : 2019-12-23 , DOI: 10.1038/s42256-019-0130-4 Wenzhi Mao , Wenze Ding , Yaoguang Xing , Haipeng Gong
|
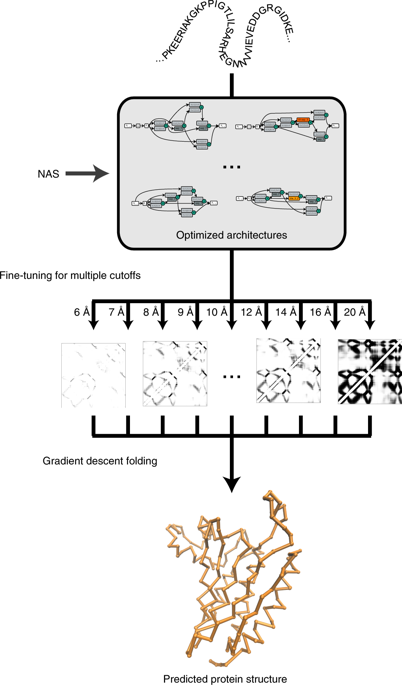
Predicting the structures of proteins from amino acid sequences is of great importance. Recently, the accuracy of de novo protein structure prediction has been substantially improved when assisted by information about the contact between residues, which is also predictable from the sequence. Here, we present a novel pipeline for rapid protein structure prediction, which consists of a residue contact predictor, AmoebaContact, and a contact-assisted folder, GDFold. Unlike mainstream contact predictors that utilize simple, regularized neural networks, AmoebaContact adopts a set of network architectures that are optimized for contact prediction through automatic searching, and it predicts contacts at a series of cutoffs. Unlike conventional contact-assisted folders that only use top-scored contact pairs, GDFold considers all residue pairs from the prediction results of AmoebaContact in a differentiable loss function and optimizes atom coordinates using the gradient descent algorithm. The combination of AmoebaContact and GDFold allows quick modelling of the protein structure with acceptable model quality.
A preprint version of the article is available at ArXiv.中文翻译:

AmoebaContact和GDFold作为快速从头进行蛋白质结构预测的管道
从氨基酸序列预测蛋白质的结构非常重要。最近,当从残基之间的接触信息中获得信息时,从头开始预测蛋白质结构的准确性已经大大提高,这也可以从序列中预测出来。在这里,我们介绍了一种用于快速蛋白质结构预测的新型管道,该管道由残基接触预测子AmoebaContact和接触辅助文件夹GDFold组成。与利用简单,规则化神经网络的主流接触预测器不同,AmoebaContact采用了一组针对通过自动搜索进行接触预测进行了优化的网络体系结构,并且它会在一系列中断时预测接触。与仅使用得分最高的联系人对的传统联系人辅助文件夹不同,GDFold考虑可变形损失函数中AmoebaContact预测结果的所有残基对,并使用梯度下降算法优化原子坐标。AmoebaContact和GDFold的组合可对蛋白质结构进行快速建模,并具有可接受的模型质量。
该文章的预印本可在ArXiv上获得。










































 京公网安备 11010802027423号
京公网安备 11010802027423号