Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
How enzyme promiscuity and horizontal gene transfer contribute to metabolic innovation.
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-01-10 , DOI: 10.1111/febs.15185 Margaret E Glasner 1 , Dat P Truong 1 , Benjamin C Morse 1
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-01-10 , DOI: 10.1111/febs.15185 Margaret E Glasner 1 , Dat P Truong 1 , Benjamin C Morse 1
Affiliation
|
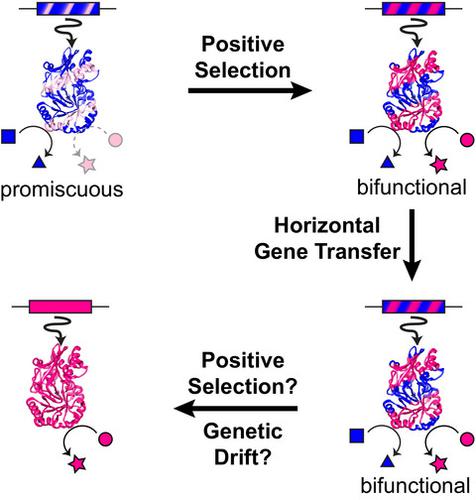
Promiscuity is the coincidental ability of an enzyme to catalyze its native reaction and additional reactions that are not biological functions in the same active site. Promiscuity plays a central role in enzyme evolution and is thus a useful property for protein and metabolic engineering. This review examines enzyme evolution holistically, beginning with evaluating biochemical support for four enzyme evolution models. As expected, there is strong biochemical support for the subfunctionalization and innovation-amplification-divergence models, in which promiscuity is a central feature. In many cases, however, enzyme evolution is more complex than the models indicate, suggesting much is yet to be learned about selective pressures on enzyme function. A complete understanding of enzyme evolution must also explain the ability of metabolic networks to integrate new enzyme activities. Hidden within metabolic networks are underground metabolic pathways constructed from promiscuous activities. We discuss efforts to determine the diversity and pervasiveness of underground metabolism. Remarkably, several studies have discovered that some metabolic defects can be repaired via multiple underground routes. In prokaryotes, metabolic innovation is driven by connecting enzymes acquired by horizontal gene transfer (HGT) into the metabolic network. Thus, we end the review by discussing how the combination of promiscuity and HGT contribute to evolution of metabolism in prokaryotes. Future studies investigating the contribution of promiscuity to enzyme and metabolic evolution will need to integrate deeper probes into the influence of evolution on protein biophysics, enzymology, and metabolism with more complex and realistic evolutionary models. ENZYMES: lactate dehydrogenase (EC 1.1.1.27), malate dehydrogenase (EC 1.1.1.37), OSBS (EC 4.2.1.113), HisA (EC 5.3.1.16), TrpF, PriA (EC 5.3.1.24), R-mandelonitrile lyase (EC 4.1.2.10), Maleylacetate reductase (EC 1.3.1.32).
中文翻译:

酶混杂和水平基因转移如何促进代谢创新。
滥交是酶催化其天然反应和在同一活性位点具有非生物学功能的其他反应的偶然能力。滥交在酶的进化中起着核心作用,因此是蛋白质和代谢工程的有用特性。这篇综述从评估四种酶进化模型的生化支持开始,全面地考察了酶的进化。不出所料,对亚功能化和创新-扩增-差异模型有强大的生化支持,其中混杂是一个主要特征。但是,在许多情况下,酶的进化要比模型所表明的更为复杂,这表明关于酶功能的选择性压力尚待研究。对酶进化的完整理解还必须解释代谢网络整合新酶活性的能力。隐藏在新陈代谢网络中的是通过混杂活动构建的地下新陈代谢途径。我们讨论确定地下代谢的多样性和普遍性的努力。值得注意的是,一些研究发现可以通过多种地下途径修复某些代谢缺陷。在原核生物中,通过将水平基因转移(HGT)获得的酶连接到代谢网络中来驱动代谢创新。因此,我们通过讨论混杂和HGT的组合如何促进原核生物代谢的演变来结束本综述。未来研究滥交对酶和代谢进化的贡献的研究将需要使用更复杂和现实的进化模型,将更深入的探针整合到进化对蛋白质生物物理学,酶学和代谢的影响中。酵素:乳酸脱氢酶(EC 1.1.1.27),苹果酸脱氢酶(EC 1.1.1.37),OSBS(EC 4.2.1.113),HisA(EC 5.3.1.16),TrpF,PriA(EC 5.3.1.24),R-扁桃腈裂解酶(EC 4.1.2.10),马来酰乙酸还原酶(EC 1.3.1.32)。
更新日期:2020-04-06
中文翻译:

酶混杂和水平基因转移如何促进代谢创新。
滥交是酶催化其天然反应和在同一活性位点具有非生物学功能的其他反应的偶然能力。滥交在酶的进化中起着核心作用,因此是蛋白质和代谢工程的有用特性。这篇综述从评估四种酶进化模型的生化支持开始,全面地考察了酶的进化。不出所料,对亚功能化和创新-扩增-差异模型有强大的生化支持,其中混杂是一个主要特征。但是,在许多情况下,酶的进化要比模型所表明的更为复杂,这表明关于酶功能的选择性压力尚待研究。对酶进化的完整理解还必须解释代谢网络整合新酶活性的能力。隐藏在新陈代谢网络中的是通过混杂活动构建的地下新陈代谢途径。我们讨论确定地下代谢的多样性和普遍性的努力。值得注意的是,一些研究发现可以通过多种地下途径修复某些代谢缺陷。在原核生物中,通过将水平基因转移(HGT)获得的酶连接到代谢网络中来驱动代谢创新。因此,我们通过讨论混杂和HGT的组合如何促进原核生物代谢的演变来结束本综述。未来研究滥交对酶和代谢进化的贡献的研究将需要使用更复杂和现实的进化模型,将更深入的探针整合到进化对蛋白质生物物理学,酶学和代谢的影响中。酵素:乳酸脱氢酶(EC 1.1.1.27),苹果酸脱氢酶(EC 1.1.1.37),OSBS(EC 4.2.1.113),HisA(EC 5.3.1.16),TrpF,PriA(EC 5.3.1.24),R-扁桃腈裂解酶(EC 4.1.2.10),马来酰乙酸还原酶(EC 1.3.1.32)。









































 京公网安备 11010802027423号
京公网安备 11010802027423号