当前位置:
X-MOL 学术
›
J. Hum. Genet.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of novel pathogenic copy number variations in Charcot-Marie-Tooth disease.
Journal of Human Genetics ( IF 2.6 ) Pub Date : 2019-12-18 , DOI: 10.1038/s10038-019-0710-5 J Mortreux 1, 2 , J Bacquet 1, 2 , A Boyer 1 , E Alazard 1 , R Bellance 3 , A G Giguet-Valard 3 , M Cerino 1, 2 , M Krahn 1, 2 , F Audic 4 , B Chabrol 4 , V Laugel 5 , J P Desvignes 2 , C Béroud 1, 2 , K Nguyen 1, 2 , A Verschueren 6 , N Lévy 1, 2 , S Attarian 6 , V Delague 2 , C Missirian 1, 2 , N Bonello-Palot 1, 2
Journal of Human Genetics ( IF 2.6 ) Pub Date : 2019-12-18 , DOI: 10.1038/s10038-019-0710-5 J Mortreux 1, 2 , J Bacquet 1, 2 , A Boyer 1 , E Alazard 1 , R Bellance 3 , A G Giguet-Valard 3 , M Cerino 1, 2 , M Krahn 1, 2 , F Audic 4 , B Chabrol 4 , V Laugel 5 , J P Desvignes 2 , C Béroud 1, 2 , K Nguyen 1, 2 , A Verschueren 6 , N Lévy 1, 2 , S Attarian 6 , V Delague 2 , C Missirian 1, 2 , N Bonello-Palot 1, 2
Affiliation
|
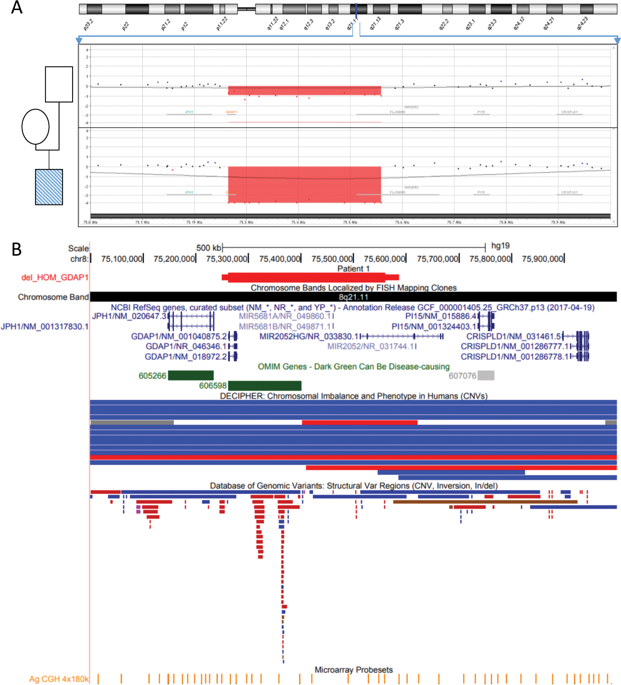
Charcot-Marie-Tooth disease (CMT) is a hereditary sensory-motor neuropathy characterized by a strong clinical and genetic heterogeneity. Over the past few years, with the occurrence of whole-exome sequencing (WES) or whole-genome sequencing (WGS), the molecular diagnosis rate has been improved by allowing the screening of more than 80 genes at one time. In CMT, except the recurrent PMP22 duplication accounting for about 60% of pathogenic variations, pathogenic copy number variations (CNVs) are rarely reported and only a few studies screening specifically CNVs have been performed. The aim of the present study was to screen for CNVs in the most prevalent genes associated with CMT in a cohort of 200 patients negative for the PMP22 duplication. CNVs were screened using the Exome Depth software on next generation sequencing (NGS) data obtained by targeted capture and sequencing of a panel of 81 CMT associated genes. Deleterious CNVs were identified in four patients (2%), in four genes: GDAP1, LRSAM1, GAN, and FGD4. All CNVs were confirmed by high-resolution oligonucleotide array Comparative Genomic Hybridization (aCGH) and/or quantitative PCR. By identifying four new CNVs in four different genes, we demonstrate that, although they are rare mutational events in CMT, CNVs might contribute significantly to mutational spectrum of Charcot-Marie-Tooth disease and should be searched in routine NGS diagnosis. This strategy increases the molecular diagnosis rate of patients with neuropathy.
中文翻译:

Charcot-Marie-Tooth 病中新型致病性拷贝数变异的鉴定。
Charcot-Marie-Tooth 病 (CMT) 是一种遗传性感觉运动神经病,其特点是具有强烈的临床和遗传异质性。过去几年,随着全外显子组测序(WES)或全基因组测序(WGS)的出现,通过允许一次筛查80多个基因,提高了分子诊断率。在 CMT 中,除了复发性 PMP22 重复约占致病变异的 60% 外,致病性拷贝数变异 (CNV) 很少报道,只有少数研究专门筛选了 CNV。本研究的目的是在 200 名 PMP22 重复阴性患者的队列中筛选与 CMT 相关的最普遍基因中的 CNV。使用 Exome Depth 软件筛选 CNV,通过对一组 81 个 CMT 相关基因进行靶向捕获和测序获得的下一代测序 (NGS) 数据。在 4 名患者 (2%) 中发现了有害的 CNV,在 4 个基因中:GDAP1、LRSAM1、GAN 和 FGD4。通过高分辨率寡核苷酸阵列比较基因组杂交 (aCGH) 和/或定量 PCR 确认所有 CNV。通过在四种不同基因中鉴定四种新的 CNV,我们证明,尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。在 4 名患者 (2%) 中发现了有害的 CNV,在 4 个基因中:GDAP1、LRSAM1、GAN 和 FGD4。通过高分辨率寡核苷酸阵列比较基因组杂交 (aCGH) 和/或定量 PCR 确认所有 CNV。通过在四种不同基因中鉴定四种新的 CNV,我们证明,尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。在 4 名患者 (2%) 中发现了有害的 CNV,在 4 个基因中:GDAP1、LRSAM1、GAN 和 FGD4。通过高分辨率寡核苷酸阵列比较基因组杂交 (aCGH) 和/或定量 PCR 确认所有 CNV。通过在四种不同基因中鉴定四种新的 CNV,我们证明,尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。
更新日期:2019-12-19
中文翻译:

Charcot-Marie-Tooth 病中新型致病性拷贝数变异的鉴定。
Charcot-Marie-Tooth 病 (CMT) 是一种遗传性感觉运动神经病,其特点是具有强烈的临床和遗传异质性。过去几年,随着全外显子组测序(WES)或全基因组测序(WGS)的出现,通过允许一次筛查80多个基因,提高了分子诊断率。在 CMT 中,除了复发性 PMP22 重复约占致病变异的 60% 外,致病性拷贝数变异 (CNV) 很少报道,只有少数研究专门筛选了 CNV。本研究的目的是在 200 名 PMP22 重复阴性患者的队列中筛选与 CMT 相关的最普遍基因中的 CNV。使用 Exome Depth 软件筛选 CNV,通过对一组 81 个 CMT 相关基因进行靶向捕获和测序获得的下一代测序 (NGS) 数据。在 4 名患者 (2%) 中发现了有害的 CNV,在 4 个基因中:GDAP1、LRSAM1、GAN 和 FGD4。通过高分辨率寡核苷酸阵列比较基因组杂交 (aCGH) 和/或定量 PCR 确认所有 CNV。通过在四种不同基因中鉴定四种新的 CNV,我们证明,尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。在 4 名患者 (2%) 中发现了有害的 CNV,在 4 个基因中:GDAP1、LRSAM1、GAN 和 FGD4。通过高分辨率寡核苷酸阵列比较基因组杂交 (aCGH) 和/或定量 PCR 确认所有 CNV。通过在四种不同基因中鉴定四种新的 CNV,我们证明,尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。在 4 名患者 (2%) 中发现了有害的 CNV,在 4 个基因中:GDAP1、LRSAM1、GAN 和 FGD4。通过高分辨率寡核苷酸阵列比较基因组杂交 (aCGH) 和/或定量 PCR 确认所有 CNV。通过在四种不同基因中鉴定四种新的 CNV,我们证明,尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。尽管它们是 CMT 中罕见的突变事件,但 CNV 可能对 Charcot-Marie-Tooth 病的突变谱有显着贡献,应在常规 NGS 诊断中进行搜索。该策略提高了神经病变患者的分子诊断率。










































 京公网安备 11010802027423号
京公网安备 11010802027423号