当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
77Se NMR Probes the Protein Environment of Selenomethionine.
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-01-07 , DOI: 10.1021/acs.jpcb.9b07466 Qingqing Chen 1 , Shiping Xu 1 , Xingyu Lu 1, 2 , Michael V Boeri 1, 3 , Yuliya Pepelyayeva 1, 4 , Elizabeth L Diaz 1 , Sunil-Datta Soni 3 , Marc Allaire 5 , Martin B Forstner 1 , Brian J Bahnson 1 , Sharon Rozovsky 1
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-01-07 , DOI: 10.1021/acs.jpcb.9b07466 Qingqing Chen 1 , Shiping Xu 1 , Xingyu Lu 1, 2 , Michael V Boeri 1, 3 , Yuliya Pepelyayeva 1, 4 , Elizabeth L Diaz 1 , Sunil-Datta Soni 3 , Marc Allaire 5 , Martin B Forstner 1 , Brian J Bahnson 1 , Sharon Rozovsky 1
Affiliation
|
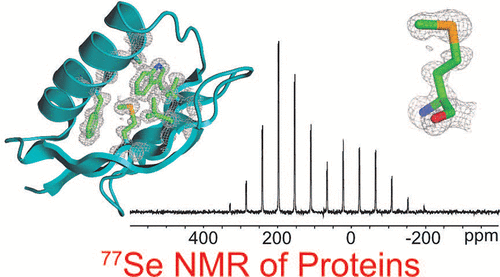
Sulfur is critical for the correct structure and proper function of proteins. Yet, lacking a sensitive enough isotope, nuclear magnetic resonance (NMR) experiments are unable to deliver for sulfur in proteins the usual wealth of chemical, dynamic, and structural information. This limitation can be circumvented by substituting sulfur with selenium, which has similar physicochemical properties and minimal impact on protein structures but possesses an NMR compatible isotope (77Se). Here we exploit the sensitivity of 77Se NMR to the nucleus' chemical milieu and use selenomethionine as a probe for its proteinaceous environment. However, such selenium NMR spectra of proteins currently resist a reliable interpretation because systematic connections between variations of system variables and changes in 77Se NMR parameters are still lacking. To start narrowing this knowledge gap, we report here on a biological 77Se magnetic resonance data bank based on a systematically designed library of GB1 variants in which a single selenomethionine was introduced at different locations within the protein. We recorded the resulting isotropic 77Se chemical shifts and relaxation times for six GB1 variants by solution-state 77Se NMR. For four of the GB1 variants we were also able to determine the chemical shift anisotropy tensor of SeM by solid-state 77Se NMR. To enable interpretation of the NMR data, the structures of five of the GB1 variants were solved by X-ray crystallography to a resolution of 1.2 Å, allowing us to unambiguously determine the conformation of the selenomethionine. Finally, we combine our solution- and solid-state NMR data with the structural information to arrive at general insights regarding the execution and interpretation of 77Se NMR experiments that exploit selenomethionine to probe proteins.
中文翻译:

77Se NMR探测硒代蛋氨酸的蛋白质环境。
硫对于蛋白质的正确结构和适当功能至关重要。但是,由于缺乏足够敏感的同位素,核磁共振(NMR)实验无法将蛋白质中的硫释放出通常具有的大量化学,动态和结构信息。可通过用硒代替硫来克服这一局限,硒具有相似的物理化学性质,对蛋白质结构的影响最小,但具有NMR兼容的同位素(77Se)。在这里,我们利用77Se NMR对原子核化学环境的敏感性,并使用硒代蛋氨酸作为其蛋白质环境的探针。但是,由于仍缺乏系统变量变化与77Se NMR参数变化之间的系统联系,因此蛋白质的此类硒NMR谱图目前尚无法可靠地解释。为了缩小这一知识鸿沟,我们在这里报告了一个生物学上的77Se磁共振数据库,该数据库基于系统设计的GB1变体库,其中在蛋白质内的不同位置引入了一个硒代蛋氨酸。我们通过溶液态77Se NMR记录了六个GB1变体的所得各向同性77Se化学位移和弛豫时间。对于GB1的四个变体,我们还能够通过固态77Se NMR确定SeM的化学位移各向异性张量。为了能够解释NMR数据,通过X射线晶体学将五个GB1变体的结构解析为1.2的分辨率,从而使我们能够明确确定硒代蛋氨酸的构象。最后,
更新日期:2020-01-07
中文翻译:

77Se NMR探测硒代蛋氨酸的蛋白质环境。
硫对于蛋白质的正确结构和适当功能至关重要。但是,由于缺乏足够敏感的同位素,核磁共振(NMR)实验无法将蛋白质中的硫释放出通常具有的大量化学,动态和结构信息。可通过用硒代替硫来克服这一局限,硒具有相似的物理化学性质,对蛋白质结构的影响最小,但具有NMR兼容的同位素(77Se)。在这里,我们利用77Se NMR对原子核化学环境的敏感性,并使用硒代蛋氨酸作为其蛋白质环境的探针。但是,由于仍缺乏系统变量变化与77Se NMR参数变化之间的系统联系,因此蛋白质的此类硒NMR谱图目前尚无法可靠地解释。为了缩小这一知识鸿沟,我们在这里报告了一个生物学上的77Se磁共振数据库,该数据库基于系统设计的GB1变体库,其中在蛋白质内的不同位置引入了一个硒代蛋氨酸。我们通过溶液态77Se NMR记录了六个GB1变体的所得各向同性77Se化学位移和弛豫时间。对于GB1的四个变体,我们还能够通过固态77Se NMR确定SeM的化学位移各向异性张量。为了能够解释NMR数据,通过X射线晶体学将五个GB1变体的结构解析为1.2的分辨率,从而使我们能够明确确定硒代蛋氨酸的构象。最后,











































 京公网安备 11010802027423号
京公网安备 11010802027423号