当前位置:
X-MOL 学术
›
Mol. Phylogenet. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Hidden cases of tRNA gene duplication and remolding in mitochondrial genomes of amphipods.
Molecular Phylogenetics and Evolution ( IF 3.6 ) Pub Date : 2019-12-15 , DOI: 10.1016/j.ympev.2019.106710 Elena V Romanova 1 , Yurij S Bukin 2 , Kirill V Mikhailov 3 , Maria D Logacheva 3 , Vladimir V Aleoshin 3 , Dmitry Yu Sherbakov 2
Molecular Phylogenetics and Evolution ( IF 3.6 ) Pub Date : 2019-12-15 , DOI: 10.1016/j.ympev.2019.106710 Elena V Romanova 1 , Yurij S Bukin 2 , Kirill V Mikhailov 3 , Maria D Logacheva 3 , Vladimir V Aleoshin 3 , Dmitry Yu Sherbakov 2
Affiliation
|
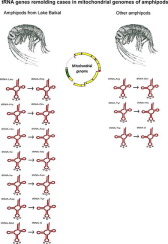
The evolution of tRNA genes in mitochondrial (mt) genomes is a complex process that includes duplications, degenerations, and transpositions, as well as a specific process of identity change through mutations in the anticodon (tRNA gene remolding or tRNA gene recruitment). Using amphipod-specific tRNA models for annotation, we show that tRNA duplications are more common in the mt genomes of amphipods than what was revealed by previous annotations. Seventeen cases of tRNA gene duplications were detected in the mt genomes of amphipods, and ten of them were tRNA genes that underwent remolding. The additional tRNA gene findings were verified using phylogenetic analysis and genetic distance analysis. The majority of remolded tRNA genes (seven out of ten cases) were found in the mt genomes of endemic amphipod species from Lake Baikal. All additional mt tRNA genes arose independently in the Baikalian amphipods, indicating the unusual plasticity of tRNA gene evolution in these species assemblages. The possible reasons for the unusual abundance of additional tRNA genes in the mt genomes of Baikalian amphipods are discussed. The amphipod-specific tRNA models developed for MiTFi refine existing predictions of tRNA genes in amphipods and reveal additional cases of duplicated tRNA genes overlooked by using less specific Metazoa-wide models. The application of these models for mt tRNA gene prediction will be useful for the correct annotation of mt genomes of amphipods and probably other crustaceans.
中文翻译:

两栖类动物线粒体基因组中tRNA基因复制和重塑的隐患。
线粒体(mt)基因组中tRNA基因的进化是一个复杂的过程,包括复制,变性和转座,以及通过反密码子突变(tRNA基因重塑或tRNA基因募集)进行身份改变的特定过程。使用两栖动物特定的tRNA模型进行注释,我们显示与两栖动物的mt基因组相比,tRNA复制比以前的注释更常见。在两栖动物的mt基因组中检测到17个tRNA基因重复的病例,其中十个是经过重塑的tRNA基因。使用系统发育分析和遗传距离分析验证了其他tRNA基因发现。在贝加尔湖特有的两栖动物物种的mt基因组中发现了大多数重塑的tRNA基因(十分之七)。所有其他mt tRNA基因均独立出现在贝加尔两栖动物纲中,表明这些物种组合中tRNA基因进化的异常可塑性。讨论了可能的原因,在贝加尔两栖动物的mt基因组中额外的tRNA基因异常丰富。为MiTFi开发的两栖动物特异性tRNA模型完善了两栖动物中tRNA基因的现有预测,并揭示了通过使用特异性较低的后生动物模型忽略了tRNA基因重复的其他情况。这些模型用于mt tRNA基因预测的应用将对正确的两栖动物和可能的其他甲壳类动物mt基因组注释有用。讨论了可能的原因,在贝加尔两栖动物的mt基因组中额外的tRNA基因异常丰富。为MiTFi开发的两栖动物特异性tRNA模型完善了两栖动物中tRNA基因的现有预测,并揭示了通过使用特异性较低的后生动物模型忽略了tRNA基因重复的其他情况。这些模型用于mt tRNA基因预测的应用将对正确的两栖动物和可能的其他甲壳类动物mt基因组注释有用。讨论了可能的原因,在贝加尔两栖动物的mt基因组中额外的tRNA基因异常丰富。为MiTFi开发的两栖动物特异性tRNA模型完善了两栖动物中tRNA基因的现有预测,并揭示了通过使用特异性较低的后生动物模型忽略了tRNA基因重复的其他情况。这些模型用于mt tRNA基因预测的应用将对正确的两栖动物和可能的其他甲壳类动物mt基因组注释有用。
更新日期:2019-12-17
中文翻译:

两栖类动物线粒体基因组中tRNA基因复制和重塑的隐患。
线粒体(mt)基因组中tRNA基因的进化是一个复杂的过程,包括复制,变性和转座,以及通过反密码子突变(tRNA基因重塑或tRNA基因募集)进行身份改变的特定过程。使用两栖动物特定的tRNA模型进行注释,我们显示与两栖动物的mt基因组相比,tRNA复制比以前的注释更常见。在两栖动物的mt基因组中检测到17个tRNA基因重复的病例,其中十个是经过重塑的tRNA基因。使用系统发育分析和遗传距离分析验证了其他tRNA基因发现。在贝加尔湖特有的两栖动物物种的mt基因组中发现了大多数重塑的tRNA基因(十分之七)。所有其他mt tRNA基因均独立出现在贝加尔两栖动物纲中,表明这些物种组合中tRNA基因进化的异常可塑性。讨论了可能的原因,在贝加尔两栖动物的mt基因组中额外的tRNA基因异常丰富。为MiTFi开发的两栖动物特异性tRNA模型完善了两栖动物中tRNA基因的现有预测,并揭示了通过使用特异性较低的后生动物模型忽略了tRNA基因重复的其他情况。这些模型用于mt tRNA基因预测的应用将对正确的两栖动物和可能的其他甲壳类动物mt基因组注释有用。讨论了可能的原因,在贝加尔两栖动物的mt基因组中额外的tRNA基因异常丰富。为MiTFi开发的两栖动物特异性tRNA模型完善了两栖动物中tRNA基因的现有预测,并揭示了通过使用特异性较低的后生动物模型忽略了tRNA基因重复的其他情况。这些模型用于mt tRNA基因预测的应用将对正确的两栖动物和可能的其他甲壳类动物mt基因组注释有用。讨论了可能的原因,在贝加尔两栖动物的mt基因组中额外的tRNA基因异常丰富。为MiTFi开发的两栖动物特异性tRNA模型完善了两栖动物中tRNA基因的现有预测,并揭示了通过使用特异性较低的后生动物模型忽略了tRNA基因重复的其他情况。这些模型用于mt tRNA基因预测的应用将对正确的两栖动物和可能的其他甲壳类动物mt基因组注释有用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号