Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Rapid bacterial detection and antibiotic susceptibility testing in whole blood using one-step, high throughput blood digital PCR.
Lab on a Chip ( IF 6.1 ) Pub Date : 2019-12-24 , DOI: 10.1039/c9lc01212e Timothy J Abram 1 , Hemanth Cherukury 2 , Chen-Yin Ou 1 , Tam Vu 3 , Michael Toledano 4 , Yiyan Li 5 , Jonathan T Grunwald 1 , Melody N Toosky 1 , Delia F Tifrea 6 , Anatoly Slepenkin 6 , Jonathan Chong 4 , Lingshun Kong 4 , Domenica Vanessa Del Pozo 4 , Kieu Thai La 4 , Louai Labanieh 4 , Jan Zimak 4 , Byron Shen 1 , Susan S Huang 7 , Enrico Gratton 8 , Ellena M Peterson 6 , Weian Zhao 9
Lab on a Chip ( IF 6.1 ) Pub Date : 2019-12-24 , DOI: 10.1039/c9lc01212e Timothy J Abram 1 , Hemanth Cherukury 2 , Chen-Yin Ou 1 , Tam Vu 3 , Michael Toledano 4 , Yiyan Li 5 , Jonathan T Grunwald 1 , Melody N Toosky 1 , Delia F Tifrea 6 , Anatoly Slepenkin 6 , Jonathan Chong 4 , Lingshun Kong 4 , Domenica Vanessa Del Pozo 4 , Kieu Thai La 4 , Louai Labanieh 4 , Jan Zimak 4 , Byron Shen 1 , Susan S Huang 7 , Enrico Gratton 8 , Ellena M Peterson 6 , Weian Zhao 9
Affiliation
|
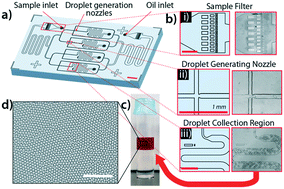
Sepsis due to antimicrobial resistant pathogens is a major health problem worldwide. The inability to rapidly detect and thus treat bacteria with appropriate agents in the early stages of infections leads to excess morbidity, mortality, and healthcare costs. Here we report a rapid diagnostic platform that integrates a novel one-step blood droplet digital PCR assay and a high throughput 3D particle counter system with potential to perform bacterial identification and antibiotic susceptibility profiling directly from whole blood specimens, without requiring culture and sample processing steps. Using CTX-M-9 family ESBLs as a model system, we demonstrated that our technology can simultaneously achieve unprecedented high sensitivity (10 CFU per ml) and rapid sample-to-answer assay time (one hour). In head-to-head studies, by contrast, real time PCR and BioRad ddPCR only exhibited a limit of detection of 1000 CFU per ml and 50-100 CFU per ml, respectively. In a blinded test inoculating clinical isolates into whole blood, we demonstrated 100% sensitivity and specificity in identifying pathogens carrying a particular resistance gene. We further demonstrated that our technology can be broadly applicable for targeted detection of a wide range of antibiotic resistant genes found in both Gram-positive (vanA, nuc, and mecA) and Gram-negative bacteria, including ESBLs (blaCTX-M-1 and blaCTX-M-2 families) and CREs (blaOXA-48 and blaKPC), as well as bacterial speciation (E. coli and Klebsiella spp.) and pan-bacterial detection, without requiring blood culture or sample processing. Our rapid diagnostic technology holds great potential in directing early, appropriate therapy and improved antibiotic stewardship in combating bloodstream infections and antibiotic resistance.
中文翻译:

使用一步,高通量血液数字PCR在全血中进行快速细菌检测和抗生素敏感性测试。
由于抗微生物病原体引起的败血症是全世界的主要健康问题。在感染的早期阶段无法快速检测细菌并用适当的试剂治疗细菌会导致较高的发病率,死亡率和医疗费用。在这里,我们报告了一个快速诊断平台,该平台集成了新颖的一步式血滴数字PCR测定法和高通量3D粒子计数器系统,可以直接从全血标本中进行细菌鉴定和抗生素敏感性分析,而无需进行培养和样品处理步骤。使用CTX-M-9系列ESBL作为模型系统,我们证明了我们的技术可以同时实现空前的高灵敏度(每毫升10 CFU)和快速的从样品到答案的测定时间(一小时)。相比之下,在面对面的研究中,实时荧光定量PCR和BioRad ddPCR的检测限分别为每毫升1000 CFU和每毫升50-100 CFU。在将临床分离株接种到全血中的盲法试验中,我们证明了在鉴定带有特定抗性基因的病原体方面具有100%的敏感性和特异性。我们进一步证明,我们的技术可广泛用于靶向检测在革兰氏阳性(vanA,nuc和mecA)和革兰氏阴性细菌(包括ESBLs(blaCTX-M-1和blaCTX-M-2家族)和CRE(blaOXA-48和blaKPC),以及细菌物种(大肠杆菌和克雷伯菌属)和泛细菌检测,而无需血液培养或样品处理。我们的快速诊断技术在早期指导方面具有巨大的潜力,
更新日期:2020-02-13
中文翻译:

使用一步,高通量血液数字PCR在全血中进行快速细菌检测和抗生素敏感性测试。
由于抗微生物病原体引起的败血症是全世界的主要健康问题。在感染的早期阶段无法快速检测细菌并用适当的试剂治疗细菌会导致较高的发病率,死亡率和医疗费用。在这里,我们报告了一个快速诊断平台,该平台集成了新颖的一步式血滴数字PCR测定法和高通量3D粒子计数器系统,可以直接从全血标本中进行细菌鉴定和抗生素敏感性分析,而无需进行培养和样品处理步骤。使用CTX-M-9系列ESBL作为模型系统,我们证明了我们的技术可以同时实现空前的高灵敏度(每毫升10 CFU)和快速的从样品到答案的测定时间(一小时)。相比之下,在面对面的研究中,实时荧光定量PCR和BioRad ddPCR的检测限分别为每毫升1000 CFU和每毫升50-100 CFU。在将临床分离株接种到全血中的盲法试验中,我们证明了在鉴定带有特定抗性基因的病原体方面具有100%的敏感性和特异性。我们进一步证明,我们的技术可广泛用于靶向检测在革兰氏阳性(vanA,nuc和mecA)和革兰氏阴性细菌(包括ESBLs(blaCTX-M-1和blaCTX-M-2家族)和CRE(blaOXA-48和blaKPC),以及细菌物种(大肠杆菌和克雷伯菌属)和泛细菌检测,而无需血液培养或样品处理。我们的快速诊断技术在早期指导方面具有巨大的潜力,











































 京公网安备 11010802027423号
京公网安备 11010802027423号