当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Saturation Mutagenesis Genome Engineering of Infective ΦX174 Bacteriophage via Unamplified Oligo Pools and Golden Gate Assembly.
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2020-01-07 , DOI: 10.1021/acssynbio.9b00411 Matthew S Faber 1 , James T Van Leuven 2, 3 , Martina M Ederer 3 , Yesol Sapozhnikov 3 , Zoë L Wilson 3 , Holly A Wichman 2, 3 , Timothy A Whitehead 4, 5 , Craig R Miller 2, 3
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2020-01-07 , DOI: 10.1021/acssynbio.9b00411 Matthew S Faber 1 , James T Van Leuven 2, 3 , Martina M Ederer 3 , Yesol Sapozhnikov 3 , Zoë L Wilson 3 , Holly A Wichman 2, 3 , Timothy A Whitehead 4, 5 , Craig R Miller 2, 3
Affiliation
|
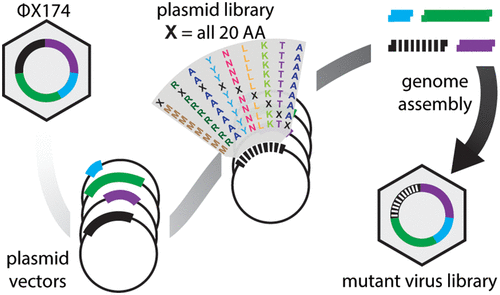
Here we present a novel protocol for the construction of saturation single-site-and massive multisite-mutant libraries of a bacteriophage. We segmented the ΦX174 genome into 14 nontoxic and nonreplicative fragments compatible with Golden Gate assembly. We next used nicking mutagenesis with oligonucleotides prepared from unamplified oligo pools with individual segments as templates to prepare near-comprehensive single-site mutagenesis libraries of genes encoding the F capsid protein (421 amino acids scanned) and G spike protein (172 amino acids scanned). Libraries possessed greater than 99% of all 11 860 programmed mutations. Golden Gate cloning was then used to assemble the complete ΦX174 mutant genome and generate libraries of infective viruses. This protocol will enable reverse genetics experiments for studying viral evolution and, with some modifications, can be applied for engineering therapeutically relevant bacteriophages with larger genomes.
中文翻译:

通过未扩增的寡核苷酸池和金门组装对感染性 ΦX174 噬菌体进行饱和诱变基因组工程。
在这里,我们提出了一种用于构建噬菌体饱和单位点和大量多位点突变体库的新方案。我们将 ΦX174 基因组分割成 14 个与 Golden Gate 组装兼容的无毒且非复制的片段。接下来,我们使用切口诱变,使用从未扩增的寡核苷酸池制备的寡核苷酸作为模板,以制备编码 F 衣壳蛋白(扫描 421 个氨基酸)和 G 刺突蛋白(扫描 172 个氨基酸)的基因的近乎全面的单位点诱变文库。文库拥有全部 11 860 个编程突变中的 99% 以上。然后使用金门克隆组装完整的 ΦX174 突变体基因组并生成感染性病毒文库。该协议将使反向遗传学实验能够用于研究病毒进化,并且经过一些修改,可以应用于具有更大基因组的工程治疗相关噬菌体。
更新日期:2020-01-07
中文翻译:

通过未扩增的寡核苷酸池和金门组装对感染性 ΦX174 噬菌体进行饱和诱变基因组工程。
在这里,我们提出了一种用于构建噬菌体饱和单位点和大量多位点突变体库的新方案。我们将 ΦX174 基因组分割成 14 个与 Golden Gate 组装兼容的无毒且非复制的片段。接下来,我们使用切口诱变,使用从未扩增的寡核苷酸池制备的寡核苷酸作为模板,以制备编码 F 衣壳蛋白(扫描 421 个氨基酸)和 G 刺突蛋白(扫描 172 个氨基酸)的基因的近乎全面的单位点诱变文库。文库拥有全部 11 860 个编程突变中的 99% 以上。然后使用金门克隆组装完整的 ΦX174 突变体基因组并生成感染性病毒文库。该协议将使反向遗传学实验能够用于研究病毒进化,并且经过一些修改,可以应用于具有更大基因组的工程治疗相关噬菌体。



























 京公网安备 11010802027423号
京公网安备 11010802027423号