当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Conditional Prediction of Ribonucleic Acid Secondary Structure Using Chemical Shifts.
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-01-08 , DOI: 10.1021/acs.jpcb.9b09814 Kexin Zhang , Aaron T. Frank
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2020-01-08 , DOI: 10.1021/acs.jpcb.9b09814 Kexin Zhang , Aaron T. Frank
|
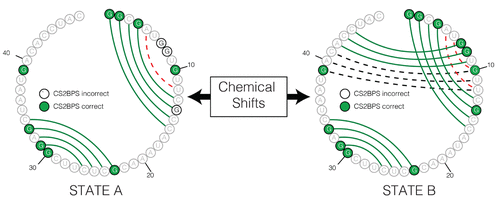
Inspired by methods that utilize chemical-mapping data to guide secondary structure prediction, we sought to develop a framework for using assigned chemical shift data to guide ribonucleic acid (RNA) secondary structure prediction. We first used machine learning to develop classifiers that predict the base-pairing status of individual residues in an RNA based on their assigned chemical shifts. Then, we used these base-pairing status predictions as restraints to guide RNA folding algorithms. Our results showed that we could recover the correct secondary fold of most of the 108 RNAs in our data set with remarkable accuracy. Finally, we tested whether we could use the base-pairing status predictions that we obtained from assigned chemical shift data to conditionally predict the secondary structure of RNA. To achieve this, we attempted to model two distinct conformational states of the microRNA-20b and the fluoride riboswitch using assigned chemical shifts that were available for both conformational states of each of these test RNAs. For both test cases, we found that by using the base-pairing status predictions that we obtained from assigned chemical shift data as folding restraints, we could generate structures that closely resembled the known structure of the two distinct states. A command-line tool for chemical shifts to base-pairing status predictions in RNA has been incorporated into our CS2Structure Git repository and can be accessed via https://github.com/atfrank/CS2Structure .
中文翻译:

使用化学位移对核糖核酸二级结构的条件预测。
受利用化学映射数据指导二级结构预测的方法的启发,我们寻求开发一种框架,用于使用分配的化学位移数据指导核糖核酸(RNA)二级结构预测。我们首先使用机器学习来开发分类器,这些分类器根据分配的化学位移预测RNA中单个残基的碱基配对状态。然后,我们使用这些碱基配对状态预测作为约束条件来指导RNA折叠算法。我们的结果表明,我们可以以极高的准确性恢复数据集中大多数108个RNA的正确二级折叠。最后,我们测试了是否可以使用从分配的化学位移数据中获得的碱基配对状态预测来有条件地预测RNA的二级结构。为了达成这个,我们试图使用分配的化学位移对microRNA-20b和氟化物核糖开关的两个不同的构象状态进行建模,这些化学位移可用于这些测试RNA的每一个的两种构象状态。对于这两个测试案例,我们发现通过使用从分配的化学位移数据获得的碱基配对状态预测作为折叠约束,我们可以生成与两种不同状态的已知结构极为相似的结构。我们已将CS2Structure Git存储库中集成了一个命令行工具,用于化学转换为RNA中的碱基配对状态预测,可通过https://github.com/atfrank/CS2Structure进行访问。
更新日期:2020-01-09
中文翻译:

使用化学位移对核糖核酸二级结构的条件预测。
受利用化学映射数据指导二级结构预测的方法的启发,我们寻求开发一种框架,用于使用分配的化学位移数据指导核糖核酸(RNA)二级结构预测。我们首先使用机器学习来开发分类器,这些分类器根据分配的化学位移预测RNA中单个残基的碱基配对状态。然后,我们使用这些碱基配对状态预测作为约束条件来指导RNA折叠算法。我们的结果表明,我们可以以极高的准确性恢复数据集中大多数108个RNA的正确二级折叠。最后,我们测试了是否可以使用从分配的化学位移数据中获得的碱基配对状态预测来有条件地预测RNA的二级结构。为了达成这个,我们试图使用分配的化学位移对microRNA-20b和氟化物核糖开关的两个不同的构象状态进行建模,这些化学位移可用于这些测试RNA的每一个的两种构象状态。对于这两个测试案例,我们发现通过使用从分配的化学位移数据获得的碱基配对状态预测作为折叠约束,我们可以生成与两种不同状态的已知结构极为相似的结构。我们已将CS2Structure Git存储库中集成了一个命令行工具,用于化学转换为RNA中的碱基配对状态预测,可通过https://github.com/atfrank/CS2Structure进行访问。











































 京公网安备 11010802027423号
京公网安备 11010802027423号