当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Modeling DNA Flexibility: Comparison of Force Fields from Atomistic to Multiscale Levels.
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2019-12-28 , DOI: 10.1021/acs.jpcb.9b09106 Vishal Minhas 1 , Tiedong Sun 1 , Alexander Mirzoev 1 , Nikolay Korolev 1 , Alexander P Lyubartsev 2 , Lars Nordenskiöld 1
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2019-12-28 , DOI: 10.1021/acs.jpcb.9b09106 Vishal Minhas 1 , Tiedong Sun 1 , Alexander Mirzoev 1 , Nikolay Korolev 1 , Alexander P Lyubartsev 2 , Lars Nordenskiöld 1
Affiliation
|
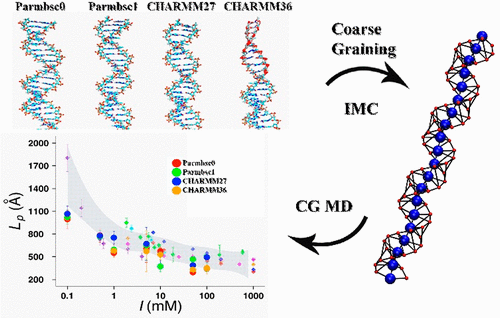
Accurate parametrization of force fields (FFs) is of ultimate importance for computer simulations to be reliable and to possess a predictive power. In this work, we analyzed, in multi-microsecond simulations of a 40-base-pair DNA fragment, the performance of four force fields, namely, the two recent major updates of CHARMM and two from the AMBER family. We focused on a description of double-helix DNA flexibility and dynamics both at atomistic and at mesoscale level in coarse-grained (CG) simulations. In addition to the traditional analysis of different base-pair and base-step parameters, we extended our analysis to investigate the ability of the force field to parametrize a CG DNA model by structure-based bottom-up coarse-graining, computing DNA persistence length as a function of ionic strength. Our simulations unambiguously showed that the CHARMM36 force field is unable to preserve DNA's structural stability at over-microsecond time scale. Both versions of the AMBER FF, parmbsc0 and parmbsc1, showed good agreement with experiment, with some bias of parmbsc0 parameters for intermediate A/B form DNA structures. The CHARMM27 force field provides stable atomistic trajectories and overall (among the considered force fields) the best fit to experimentally determined DNA flexibility parameters both at atomistic and at mesoscale level.
中文翻译:

DNA灵活性建模:从原子水平到多尺度水平的力场比较。
力场(FFs)的精确参数化对于计算机模拟的可靠性和预测能力至关重要。在这项工作中,我们在一个40个碱基对的DNA片段的多微秒模拟中,分析了四个力场的性能,即CHARMM的两个最近的重大更新以及AMBER家族的两个。我们专注于在粗粒度(CG)模拟中在原子级和中尺度水平上双螺旋DNA的灵活性和动力学的描述。除了对不同碱基对和碱基步长参数的传统分析之外,我们还扩展了分析范围,以研究力场通过基于结构的自下而上的粗粒度计算CG DNA模型的能力,计算DNA持久长度作为离子强度的函数。我们的模拟明确表明,CHARMM36力场无法在超过微秒的时间内保持DNA的结构稳定性。AMBER FF的两个版本parmbsc0和parmbsc1与实验显示出很好的一致性,其中parmbsc0参数对于中间A / B形式的DNA结构有些偏向。CHARMM27力场提供了稳定的原子轨迹,并且总体上(在考虑的力场中)最适合以原子级和中尺度水平通过实验确定的DNA柔性参数。
更新日期:2019-12-29
中文翻译:

DNA灵活性建模:从原子水平到多尺度水平的力场比较。
力场(FFs)的精确参数化对于计算机模拟的可靠性和预测能力至关重要。在这项工作中,我们在一个40个碱基对的DNA片段的多微秒模拟中,分析了四个力场的性能,即CHARMM的两个最近的重大更新以及AMBER家族的两个。我们专注于在粗粒度(CG)模拟中在原子级和中尺度水平上双螺旋DNA的灵活性和动力学的描述。除了对不同碱基对和碱基步长参数的传统分析之外,我们还扩展了分析范围,以研究力场通过基于结构的自下而上的粗粒度计算CG DNA模型的能力,计算DNA持久长度作为离子强度的函数。我们的模拟明确表明,CHARMM36力场无法在超过微秒的时间内保持DNA的结构稳定性。AMBER FF的两个版本parmbsc0和parmbsc1与实验显示出很好的一致性,其中parmbsc0参数对于中间A / B形式的DNA结构有些偏向。CHARMM27力场提供了稳定的原子轨迹,并且总体上(在考虑的力场中)最适合以原子级和中尺度水平通过实验确定的DNA柔性参数。











































 京公网安备 11010802027423号
京公网安备 11010802027423号