Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Novel copper-containing membrane monooxygenases (CuMMOs) encoded by alkane-utilizing Betaproteobacteria.
The ISME Journal ( IF 10.8 ) Pub Date : 2019-12-03 , DOI: 10.1038/s41396-019-0561-2 Fauziah F Rochman 1 , Miye Kwon 2 , Roshan Khadka 1 , Ivica Tamas 1, 3 , Azriel Abraham Lopez-Jauregui 1, 4 , Andriy Sheremet 1 , Angela V Smirnova 1 , Rex R Malmstrom 5 , Sukhwan Yoon 2 , Tanja Woyke 5 , Peter F Dunfield 1 , Tobin J Verbeke 1
The ISME Journal ( IF 10.8 ) Pub Date : 2019-12-03 , DOI: 10.1038/s41396-019-0561-2 Fauziah F Rochman 1 , Miye Kwon 2 , Roshan Khadka 1 , Ivica Tamas 1, 3 , Azriel Abraham Lopez-Jauregui 1, 4 , Andriy Sheremet 1 , Angela V Smirnova 1 , Rex R Malmstrom 5 , Sukhwan Yoon 2 , Tanja Woyke 5 , Peter F Dunfield 1 , Tobin J Verbeke 1
Affiliation
|
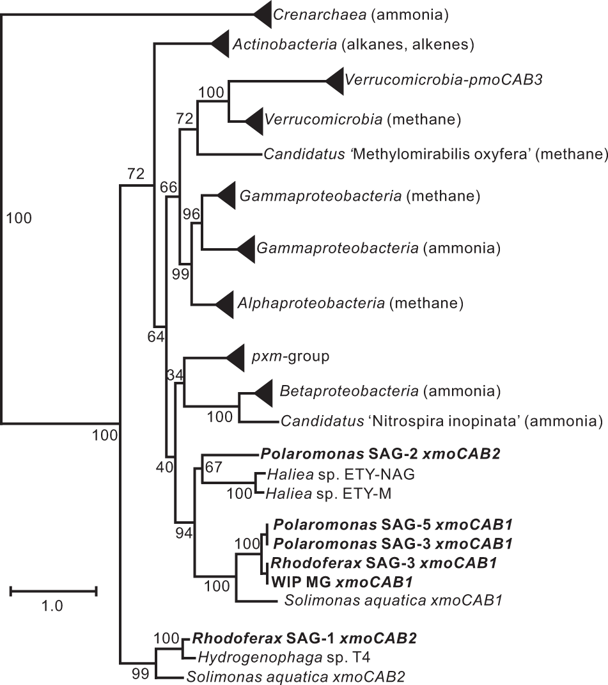
Copper-containing membrane monooxygenases (CuMMOs) are encoded by xmoCAB(D) gene clusters and catalyze the oxidation of methane, ammonia, or some short-chain alkanes and alkenes. In a metagenome constructed from an oilsands tailings pond we detected an xmoCABD gene cluster with <59% derived protein sequence identity to genes from known bacteria. Stable isotope probing experiments combined with a specific xmoA qPCR assay demonstrated that the bacteria possessing these genes were incapable of methane assimilation, but did grow on ethane and propane. Single-cell amplified genomes (SAGs) from propane-enriched samples were screened with the specific PCR assay to identify bacteria possessing the target gene cluster. Multiple SAGs of Betaproteobacteria belonging to the genera Rhodoferax and Polaromonas possessed homologues of the metagenomic xmoCABD gene cluster. Unexpectedly, each of these two genera also possessed other xmoCABD paralogs, representing two additional lineages in phylogenetic analyses. Metabolic reconstructions from SAGs predicted that neither bacterium encoded enzymes with the potential to support catabolic methane or ammonia oxidation, but that both were capable of higher n-alkane degradation. The involvement of the encoded CuMMOs in alkane oxidation was further suggested by reverse transcription PCR analyses, which detected elevated transcription of the xmoA genes upon enrichment of water samples with propane as the sole energy source. Enrichments, isotope incorporation studies, genome reconstructions, and gene expression studies therefore all agreed that the unknown xmoCABD operons did not encode methane or ammonia monooxygenases, but rather n-alkane monooxygenases. This study broadens the known diversity of CuMMOs and identifies these enzymes in non-nitrifying Betaproteobacteria.
中文翻译:

由利用烷烃的 Betaproteobacteria 编码的新型含铜膜单加氧酶 (CuMMOs)。
含铜膜单加氧酶 (CuMMOs) 由 xmoCAB(D) 基因簇编码,催化甲烷、氨或一些短链烷烃和烯烃的氧化。在由油砂尾矿池构建的宏基因组中,我们检测到 xmoCABD 基因簇与已知细菌基因的衍生蛋白质序列同一性<59%。稳定同位素探测实验结合特定的 xmoA qPCR 测定表明,拥有这些基因的细菌不能吸收甲烷,但确实在乙烷和丙烷上生长。用特定的 PCR 测定法筛选来自富含丙烷的样品的单细胞扩增基因组 (SAG),以鉴定具有靶基因簇的细菌。属于 Rhodoferax 和 Polaromonas 属的 Betaproteobacteria 的多个 SAG 具有宏基因组 xmoCABD 基因簇的同源物。出乎意料的是,这两个属中的每一个还拥有其他 xmoCABD 旁系同源物,代表系统发育分析中的两个额外谱系。来自 SAG 的代谢重建预测,两种细菌都没有编码具有支持分解代谢甲烷或氨氧化潜力的酶,但两者都能够进行更高的正烷烃降解。逆转录 PCR 分析进一步表明编码的 CuMMOs 参与烷烃氧化,该分析检测到以丙烷为唯一能源的水样富集后 xmoA 基因的转录升高。富集、同位素掺入研究、基因组重建、因此,基因表达研究都同意未知的 xmoCABD 操纵子不编码甲烷或氨单加氧酶,而是编码正烷烃单加氧酶。这项研究拓宽了 CuMMO 的已知多样性,并在非硝化 Betaproteobacteria 中鉴定了这些酶。
更新日期:2020-01-17
中文翻译:

由利用烷烃的 Betaproteobacteria 编码的新型含铜膜单加氧酶 (CuMMOs)。
含铜膜单加氧酶 (CuMMOs) 由 xmoCAB(D) 基因簇编码,催化甲烷、氨或一些短链烷烃和烯烃的氧化。在由油砂尾矿池构建的宏基因组中,我们检测到 xmoCABD 基因簇与已知细菌基因的衍生蛋白质序列同一性<59%。稳定同位素探测实验结合特定的 xmoA qPCR 测定表明,拥有这些基因的细菌不能吸收甲烷,但确实在乙烷和丙烷上生长。用特定的 PCR 测定法筛选来自富含丙烷的样品的单细胞扩增基因组 (SAG),以鉴定具有靶基因簇的细菌。属于 Rhodoferax 和 Polaromonas 属的 Betaproteobacteria 的多个 SAG 具有宏基因组 xmoCABD 基因簇的同源物。出乎意料的是,这两个属中的每一个还拥有其他 xmoCABD 旁系同源物,代表系统发育分析中的两个额外谱系。来自 SAG 的代谢重建预测,两种细菌都没有编码具有支持分解代谢甲烷或氨氧化潜力的酶,但两者都能够进行更高的正烷烃降解。逆转录 PCR 分析进一步表明编码的 CuMMOs 参与烷烃氧化,该分析检测到以丙烷为唯一能源的水样富集后 xmoA 基因的转录升高。富集、同位素掺入研究、基因组重建、因此,基因表达研究都同意未知的 xmoCABD 操纵子不编码甲烷或氨单加氧酶,而是编码正烷烃单加氧酶。这项研究拓宽了 CuMMO 的已知多样性,并在非硝化 Betaproteobacteria 中鉴定了这些酶。











































 京公网安备 11010802027423号
京公网安备 11010802027423号