当前位置:
X-MOL 学术
›
JAMA Dermatol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Keratinocytic Skin Cancer Detection on the Face Using Region-Based Convolutional Neural Network.
JAMA Dermatology ( IF 11.5 ) Pub Date : 2019-12-04 , DOI: 10.1001/jamadermatol.2019.3807 Seung Seog Han 1 , Ik Jun Moon 2 , Woohyung Lim 3 , In Suck Suh 4 , Sam Yong Lee 5 , Jung-Im Na 6 , Seong Hwan Kim 4 , Sung Eun Chang 7
JAMA Dermatology ( IF 11.5 ) Pub Date : 2019-12-04 , DOI: 10.1001/jamadermatol.2019.3807 Seung Seog Han 1 , Ik Jun Moon 2 , Woohyung Lim 3 , In Suck Suh 4 , Sam Yong Lee 5 , Jung-Im Na 6 , Seong Hwan Kim 4 , Sung Eun Chang 7
Affiliation
|
Importance
Detection of cutaneous cancer on the face using deep-learning algorithms has been challenging because various anatomic structures create curves and shades that confuse the algorithm and can potentially lead to false-positive results.
Objective
To evaluate whether an algorithm can automatically locate suspected areas and predict the probability of a lesion being malignant.
Design, Setting, and Participants
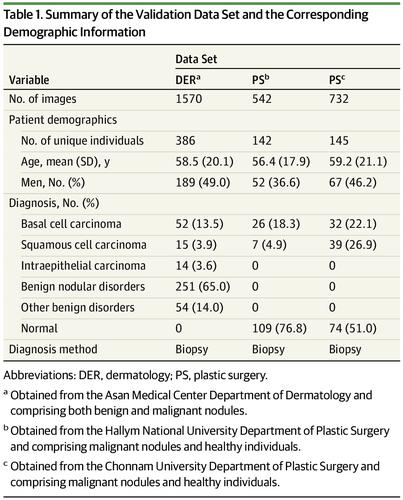
Region-based convolutional neural network technology was used to create 924 538 possible lesions by extracting nodular benign lesions from 182 348 clinical photographs. After manually or automatically annotating these possible lesions based on image findings, convolutional neural networks were trained with 1 106 886 image crops to locate and diagnose cancer. Validation data sets (2844 images from 673 patients; mean [SD] age, 58.2 [19.9] years; 308 men [45.8%]; 185 patients with malignant tumors, 305 with benign tumors, and 183 free of tumor) were obtained from 3 hospitals between January 1, 2010, and September 30, 2018.
Main Outcomes and Measures
The area under the receiver operating characteristic curve, F1 score (mean of precision and recall; range, 0.000-1.000), and Youden index score (sensitivity + specificity -1; 0%-100%) were used to compare the performance of the algorithm with that of the participants.
Results
The algorithm analyzed a mean (SD) of 4.2 (2.4) photographs per patient and reported the malignancy score according to the highest malignancy output. The area under the receiver operating characteristic curve for the validation data set (673 patients) was 0.910. At a high-sensitivity cutoff threshold, the sensitivity and specificity of the model with the 673 patients were 76.8% and 90.6%, respectively. With the test partition (325 images; 80 patients), the performance of the algorithm was compared with the performance of 13 board-certified dermatologists, 34 dermatology residents, 20 nondermatologic physicians, and 52 members of the general public with no medical background. When the disease screening performance was evaluated at high sensitivity areas using the F1 score and Youden index score, the algorithm showed a higher F1 score (0.831 vs 0.653 [0.126], P < .001) and Youden index score (0.675 vs 0.417 [0.124], P < .001) than that of nondermatologic physicians. The accuracy of the algorithm was comparable with that of dermatologists (F1 score, 0.831 vs 0.835 [0.040]; Youden index score, 0.675 vs 0.671 [0.100]).
Conclusions and Relevance
The results of the study suggest that the algorithm could localize and diagnose skin cancer without preselection of suspicious lesions by dermatologists.
中文翻译:

使用基于区域的卷积神经网络检测面部角化细胞性皮肤癌。
使用深度学习算法检测面部皮肤癌的重要性一直很具有挑战性,因为各种解剖结构会产生曲线和阴影,从而使算法感到困惑,并有可能导致假阳性结果。目的评估算法是否可以自动定位可疑区域并预测病变为恶性的可能性。设计,设置和参与者基于区域的卷积神经网络技术用于通过从182348张临床照片中提取结节性良性病变来创建924538个可能的病变。在基于图像发现手动或自动注释这些可能的病变后,使用1 106 886个图像作物训练了卷积神经网络,以定位和诊断癌症。验证数据集(来自673例患者的2844张图像;平均[SD]年龄为58.2 [19。9]年;308名男性[45.8%];在2010年1月1日至2018年9月30日之间从3所医院中获得185例恶性肿瘤,305例良性肿瘤和183例无肿瘤。主要结果和措施受试者工作特征曲线下的面积,F1评分(精确度和召回率的平均值;范围为0.000-1.000)和Youden指数得分(敏感性+特异性-1; 0%-100%)用于比较该算法与参与者的性能。结果该算法对每位患者的4.2(2.4)张照片进行了平均(SD)分析,并根据最高恶性肿瘤输出报告了恶性评分。验证数据集(673例患者)的接收器工作特征曲线下方的面积为0.910。在高灵敏度截止阈值下,该模型对673名患者的敏感性和特异性分别为76.8%和90.6%。使用测试分区(325张图像; 80位患者),将算法的性能与13位经董事会认证的皮肤科医生,34位皮肤科医师,20位非皮肤科医师以及52位无医学背景的普通公众的性能进行了比较。当使用F1得分和Youden指数得分在高灵敏度区域评估疾病筛查性能时,该算法显示出更高的F1得分(0.831 vs 0.653 [0.126],P <.001)和Youden指数得分(0.675 vs 0.417 [0.124]) ],P <.001)比非皮肤科医师大。该算法的准确性与皮肤科医生相当(F1评分为0.831 vs 0.835 [0.040]; Youden指数评分为0.675 vs 0.671 [0.100])。
更新日期:2020-01-08
中文翻译:

使用基于区域的卷积神经网络检测面部角化细胞性皮肤癌。
使用深度学习算法检测面部皮肤癌的重要性一直很具有挑战性,因为各种解剖结构会产生曲线和阴影,从而使算法感到困惑,并有可能导致假阳性结果。目的评估算法是否可以自动定位可疑区域并预测病变为恶性的可能性。设计,设置和参与者基于区域的卷积神经网络技术用于通过从182348张临床照片中提取结节性良性病变来创建924538个可能的病变。在基于图像发现手动或自动注释这些可能的病变后,使用1 106 886个图像作物训练了卷积神经网络,以定位和诊断癌症。验证数据集(来自673例患者的2844张图像;平均[SD]年龄为58.2 [19。9]年;308名男性[45.8%];在2010年1月1日至2018年9月30日之间从3所医院中获得185例恶性肿瘤,305例良性肿瘤和183例无肿瘤。主要结果和措施受试者工作特征曲线下的面积,F1评分(精确度和召回率的平均值;范围为0.000-1.000)和Youden指数得分(敏感性+特异性-1; 0%-100%)用于比较该算法与参与者的性能。结果该算法对每位患者的4.2(2.4)张照片进行了平均(SD)分析,并根据最高恶性肿瘤输出报告了恶性评分。验证数据集(673例患者)的接收器工作特征曲线下方的面积为0.910。在高灵敏度截止阈值下,该模型对673名患者的敏感性和特异性分别为76.8%和90.6%。使用测试分区(325张图像; 80位患者),将算法的性能与13位经董事会认证的皮肤科医生,34位皮肤科医师,20位非皮肤科医师以及52位无医学背景的普通公众的性能进行了比较。当使用F1得分和Youden指数得分在高灵敏度区域评估疾病筛查性能时,该算法显示出更高的F1得分(0.831 vs 0.653 [0.126],P <.001)和Youden指数得分(0.675 vs 0.417 [0.124]) ],P <.001)比非皮肤科医师大。该算法的准确性与皮肤科医生相当(F1评分为0.831 vs 0.835 [0.040]; Youden指数评分为0.675 vs 0.671 [0.100])。











































 京公网安备 11010802027423号
京公网安备 11010802027423号