当前位置:
X-MOL 学术
›
Nat. Biotechnol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Deciphering eukaryotic gene-regulatory logic with 100 million random promoters.
Nature Biotechnology ( IF 33.1 ) Pub Date : 2019-12-02 , DOI: 10.1038/s41587-019-0315-8 Carl G de Boer 1 , Eeshit Dhaval Vaishnav 1, 2 , Ronen Sadeh 3 , Esteban Luis Abeyta 4 , Nir Friedman 1, 3 , Aviv Regev 1, 2
Nature Biotechnology ( IF 33.1 ) Pub Date : 2019-12-02 , DOI: 10.1038/s41587-019-0315-8 Carl G de Boer 1 , Eeshit Dhaval Vaishnav 1, 2 , Ronen Sadeh 3 , Esteban Luis Abeyta 4 , Nir Friedman 1, 3 , Aviv Regev 1, 2
Affiliation
|
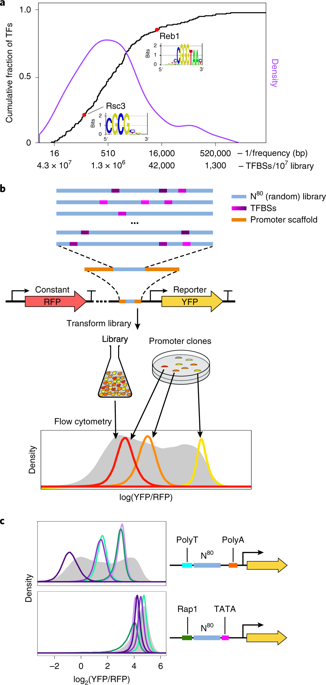
How transcription factors (TFs) interpret cis-regulatory DNA sequence to control gene expression remains unclear, largely because past studies using native and engineered sequences had insufficient scale. Here, we measure the expression output of >100 million synthetic yeast promoter sequences that are fully random. These sequences yield diverse, reproducible expression levels that can be explained by their chance inclusion of functional TF binding sites. We use machine learning to build interpretable models of transcriptional regulation that predict ~94% of the expression driven from independent test promoters and ~89% of the expression driven from native yeast promoter fragments. These models allow us to characterize each TF's specificity, activity and interactions with chromatin. TF activity depends on binding-site strand, position, DNA helical face and chromatin context. Notably, expression level is influenced by weak regulatory interactions, which confound designed-sequence studies. Our analyses show that massive-throughput assays of fully random DNA can provide the big data necessary to develop complex, predictive models of gene regulation.
中文翻译:

用 1 亿个随机启动子破译真核基因调控逻辑。
转录因子 (TF) 如何解释顺式调控 DNA 序列以控制基因表达仍不清楚,主要是因为过去使用天然和工程序列的研究规模不足。在这里,我们测量了超过 1 亿个完全随机的合成酵母启动子序列的表达输出。这些序列产生不同的、可重复的表达水平,这可以通过它们偶然包含功能性 TF 结合位点来解释。我们使用机器学习来构建可解释的转录调控模型,预测约 94% 的表达来自独立测试启动子,约 89% 的表达来自天然酵母启动子片段。这些模型使我们能够表征每个 TF 的特异性、活性和与染色质的相互作用。TF 活性取决于结合位点链、位置、DNA 螺旋面和染色质背景。值得注意的是,表达水平受弱调节相互作用的影响,这混淆了设计序列研究。我们的分析表明,完全随机 DNA 的大通量分析可以提供开发复杂的基因调控预测模型所需的大数据。
更新日期:2019-12-02
中文翻译:

用 1 亿个随机启动子破译真核基因调控逻辑。
转录因子 (TF) 如何解释顺式调控 DNA 序列以控制基因表达仍不清楚,主要是因为过去使用天然和工程序列的研究规模不足。在这里,我们测量了超过 1 亿个完全随机的合成酵母启动子序列的表达输出。这些序列产生不同的、可重复的表达水平,这可以通过它们偶然包含功能性 TF 结合位点来解释。我们使用机器学习来构建可解释的转录调控模型,预测约 94% 的表达来自独立测试启动子,约 89% 的表达来自天然酵母启动子片段。这些模型使我们能够表征每个 TF 的特异性、活性和与染色质的相互作用。TF 活性取决于结合位点链、位置、DNA 螺旋面和染色质背景。值得注意的是,表达水平受弱调节相互作用的影响,这混淆了设计序列研究。我们的分析表明,完全随机 DNA 的大通量分析可以提供开发复杂的基因调控预测模型所需的大数据。











































 京公网安备 11010802027423号
京公网安备 11010802027423号