当前位置:
X-MOL 学术
›
npj Genom. Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Re-annotation of 191 developmental and epileptic encephalopathy-associated genes unmasks de novo variants in SCN1A.
npj Genomic Medicine ( IF 4.7 ) Pub Date : 2019-12-02 , DOI: 10.1038/s41525-019-0106-7 Charles A Steward 1, 2 , Jolien Roovers 3, 4 , Marie-Marthe Suner 2, 5 , Jose M Gonzalez 2, 5 , Barbara Uszczynska-Ratajczak 6, 7, 8 , Dmitri Pervouchine 9 , Stephen Fitzgerald 2 , Margarida Viola 3, 4 , Hannah Stamberger 3, 4, 10 , Fadi F Hamdan 11 , Berten Ceulemans 12 , Patricia Leroy 13 , Caroline Nava 14, 15 , Anne Lepine 16 , Electra Tapanari 2, 5 , Don Keiller 17 , Stephen Abbs 18 , Alba Sanchis-Juan 19 , Detelina Grozeva 20 , Anthony S Rogers 1 , Mark Diekhans 21 , Roderic Guigó 6, 7 , Robert Petryszak 5 , Berge A Minassian 22, 23 , Gianpiero Cavalleri 24 , Dimitrios Vitsios 25 , Slavé Petrovski 25 , Jennifer Harrow 2, 5, 26 , Paul Flicek 5 , F Lucy Raymond 20 , Nicholas J Lench 1, 27 , Peter De Jonghe 3, 4, 10 , Jonathan M Mudge 2, 5 , Sarah Weckhuysen 3, 4, 10 , Sanjay M Sisodiya 28, 29 , Adam Frankish 2, 5
npj Genomic Medicine ( IF 4.7 ) Pub Date : 2019-12-02 , DOI: 10.1038/s41525-019-0106-7 Charles A Steward 1, 2 , Jolien Roovers 3, 4 , Marie-Marthe Suner 2, 5 , Jose M Gonzalez 2, 5 , Barbara Uszczynska-Ratajczak 6, 7, 8 , Dmitri Pervouchine 9 , Stephen Fitzgerald 2 , Margarida Viola 3, 4 , Hannah Stamberger 3, 4, 10 , Fadi F Hamdan 11 , Berten Ceulemans 12 , Patricia Leroy 13 , Caroline Nava 14, 15 , Anne Lepine 16 , Electra Tapanari 2, 5 , Don Keiller 17 , Stephen Abbs 18 , Alba Sanchis-Juan 19 , Detelina Grozeva 20 , Anthony S Rogers 1 , Mark Diekhans 21 , Roderic Guigó 6, 7 , Robert Petryszak 5 , Berge A Minassian 22, 23 , Gianpiero Cavalleri 24 , Dimitrios Vitsios 25 , Slavé Petrovski 25 , Jennifer Harrow 2, 5, 26 , Paul Flicek 5 , F Lucy Raymond 20 , Nicholas J Lench 1, 27 , Peter De Jonghe 3, 4, 10 , Jonathan M Mudge 2, 5 , Sarah Weckhuysen 3, 4, 10 , Sanjay M Sisodiya 28, 29 , Adam Frankish 2, 5
Affiliation
|
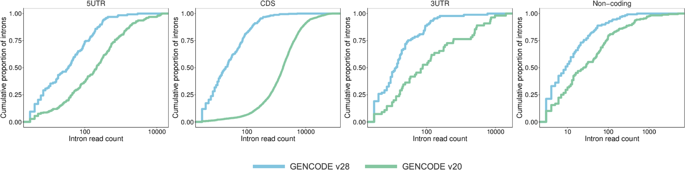
The developmental and epileptic encephalopathies (DEE) are a group of rare, severe neurodevelopmental disorders, where even the most thorough sequencing studies leave 60-65% of patients without a molecular diagnosis. Here, we explore the incompleteness of transcript models used for exome and genome analysis as one potential explanation for a lack of current diagnoses. Therefore, we have updated the GENCODE gene annotation for 191 epilepsy-associated genes, using human brain-derived transcriptomic libraries and other data to build 3,550 putative transcript models. Our annotations increase the transcriptional 'footprint' of these genes by over 674 kb. Using SCN1A as a case study, due to its close phenotype/genotype correlation with Dravet syndrome, we screened 122 people with Dravet syndrome or a similar phenotype with a panel of exon sequences representing eight established genes and identified two de novo SCN1A variants that now - through improved gene annotation - are ascribed to residing among our exons. These two (from 122 screened people, 1.6%) molecular diagnoses carry significant clinical implications. Furthermore, we identified a previously classified SCN1A intronic Dravet syndrome-associated variant that now lies within a deeply conserved exon. Our findings illustrate the potential gains of thorough gene annotation in improving diagnostic yields for genetic disorders.
中文翻译:

191个发育性和癫痫性脑病相关基因的重新注释揭示了SCN1A中的从头变异。
发育性和癫痫性脑病(DEE)是一组罕见的严重神经发育障碍,即使最彻底的测序研究也使60-65%的患者无法进行分子诊断。在这里,我们探索用于外显子组和基因组分析的转录本模型的不完整,作为目前缺乏诊断的一种潜在解释。因此,我们使用人脑衍生的转录组文库和其他数据,为191个癫痫相关基因更新了GENCODE基因注释,以建立3,550个假定的转录本模型。我们的注释使这些基因的转录“足迹”增加了674 kb以上。以SCN1A为例,由于其表型/基因型与Dravet综合征密切相关,我们筛选了122名患有Dravet综合征或类似表型的人,这些人的外显子序列代表了八个已建立的基因,并鉴定了两个从头开始的SCN1A变体,这些变体现在通过改进的基因注释而归属于我们的外显子。这两个分子(来自122位筛查者中,占1.6%)的分子诊断具有重要的临床意义。此外,我们确定了先前分类的SCN1A内含子Dravet综合征相关变体,现在位于一个高度保守的外显子中。我们的发现说明了彻底的基因注释在提高遗传疾病诊断率方面的潜在收益。6%)的分子诊断具有重大的临床意义。此外,我们确定了先前分类的SCN1A内含子Dravet综合征相关变体,现在位于一个高度保守的外显子中。我们的发现说明了彻底的基因注释在提高遗传疾病诊断率方面的潜在收益。6%)的分子诊断具有重大的临床意义。此外,我们确定了先前分类的SCN1A内含子Dravet综合征相关变体,现在位于一个高度保守的外显子中。我们的发现说明了彻底的基因注释在提高遗传疾病诊断率方面的潜在收益。
更新日期:2019-12-02
中文翻译:

191个发育性和癫痫性脑病相关基因的重新注释揭示了SCN1A中的从头变异。
发育性和癫痫性脑病(DEE)是一组罕见的严重神经发育障碍,即使最彻底的测序研究也使60-65%的患者无法进行分子诊断。在这里,我们探索用于外显子组和基因组分析的转录本模型的不完整,作为目前缺乏诊断的一种潜在解释。因此,我们使用人脑衍生的转录组文库和其他数据,为191个癫痫相关基因更新了GENCODE基因注释,以建立3,550个假定的转录本模型。我们的注释使这些基因的转录“足迹”增加了674 kb以上。以SCN1A为例,由于其表型/基因型与Dravet综合征密切相关,我们筛选了122名患有Dravet综合征或类似表型的人,这些人的外显子序列代表了八个已建立的基因,并鉴定了两个从头开始的SCN1A变体,这些变体现在通过改进的基因注释而归属于我们的外显子。这两个分子(来自122位筛查者中,占1.6%)的分子诊断具有重要的临床意义。此外,我们确定了先前分类的SCN1A内含子Dravet综合征相关变体,现在位于一个高度保守的外显子中。我们的发现说明了彻底的基因注释在提高遗传疾病诊断率方面的潜在收益。6%)的分子诊断具有重大的临床意义。此外,我们确定了先前分类的SCN1A内含子Dravet综合征相关变体,现在位于一个高度保守的外显子中。我们的发现说明了彻底的基因注释在提高遗传疾病诊断率方面的潜在收益。6%)的分子诊断具有重大的临床意义。此外,我们确定了先前分类的SCN1A内含子Dravet综合征相关变体,现在位于一个高度保守的外显子中。我们的发现说明了彻底的基因注释在提高遗传疾病诊断率方面的潜在收益。









































 京公网安备 11010802027423号
京公网安备 11010802027423号