European Journal of Human Genetics ( IF 3.7 ) Pub Date : 2019-11-29 , DOI: 10.1038/s41431-019-0551-x Massimiliano Cocca 1 , Caterina Barbieri 2 , Maria Pina Concas 1 , Antonietta Robino 1 , Marco Brumat 3 , Ilaria Gandin 3 , Matteo Trudu 4 , Cinzia Felicita Sala 2 , Dragana Vuckovic 5 , Giorgia Girotto 1, 3 , Giuseppe Matullo 6, 7 , Ozren Polasek 8 , Ivana Kolčić 8 , Paolo Gasparini 1, 3 , Nicole Soranzo 5 , Daniela Toniolo 2 , Massimo Mezzavilla 1
|
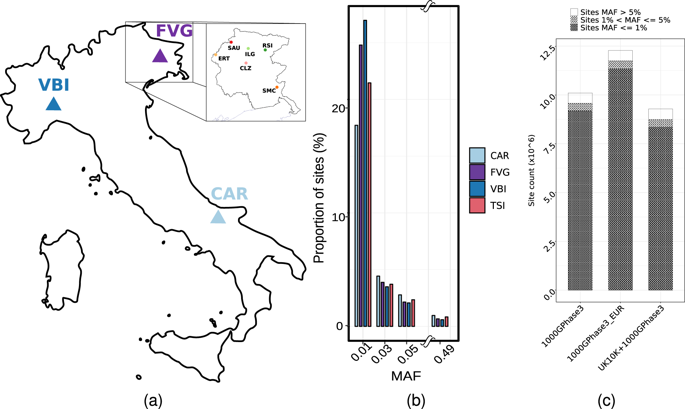
The genomic variation of the Italian peninsula populations is currently under characterised: the only Italian whole-genome reference is represented by the Tuscans from the 1000 Genome Project. To address this issue, we sequenced a total of 947 Italian samples from three different geographical areas. First, we defined a new Italian Genome Reference Panel (IGRP1.0) for imputation, which improved imputation accuracy, especially for rare variants, and we tested it by GWAS analysis on red blood traits. Furthermore, we extended the catalogue of genetic variation investigating the level of population structure, the pattern of natural selection, the distribution of deleterious variants and occurrence of human knockouts (HKOs). Overall the results demonstrate a high level of genomic differentiation between cohorts, different signatures of natural selection and a distinctive distribution of deleterious variants and HKOs, confirming the necessity of distinct genome references for the Italian population.
中文翻译:

通过全基因组测序鸟瞰意大利基因组变异。
意大利半岛种群的基因组变异目前正在表征:唯一的意大利全基因组参考由来自 1000 基因组计划的托斯卡纳人代表。为了解决这个问题,我们对来自三个不同地理区域的总共 947 个意大利样本进行了测序。首先,我们定义了一个新的意大利基因组参考面板 (IGRP1.0) 用于插补,这提高了插补的准确性,尤其是对于罕见变异,我们通过对红血性状的 GWAS 分析对其进行了测试。此外,我们扩展了遗传变异目录,调查种群结构水平、自然选择模式、有害变异的分布和人类基因敲除 (HKO) 的发生。总体而言,结果表明队列之间存在高水平的基因组分化,









































 京公网安备 11010802027423号
京公网安备 11010802027423号