当前位置:
X-MOL 学术
›
Chem. Res. Toxicol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Prediction of Adverse Drug Reactions by Combining Biomedical Tripartite Network and Graph Representation Model.
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2019-12-13 , DOI: 10.1021/acs.chemrestox.9b00238 Rui Xue 1 , Jie Liao 1 , Xin Shao 1 , Ke Han 1 , Jingbo Long 1 , Li Shao 2 , Ni Ai 1 , Xiaohui Fan 1
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2019-12-13 , DOI: 10.1021/acs.chemrestox.9b00238 Rui Xue 1 , Jie Liao 1 , Xin Shao 1 , Ke Han 1 , Jingbo Long 1 , Li Shao 2 , Ni Ai 1 , Xiaohui Fan 1
Affiliation
|
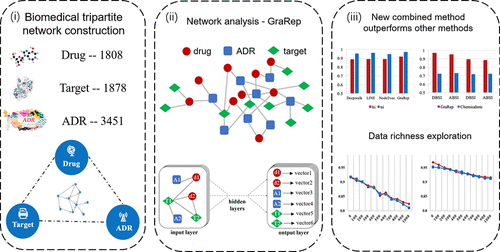
As one of the primary contributors to high clinical attrition rates of drugs, toxicity evaluation is of critical significance to new drug discovery. Unsurprisingly, a vast number of computational methods have been developed at various stages of development pipeline to evaluate potential adverse drug reactions (ADRs). Despite previous success of these methods on individual ADR or certain drug family, there are great challenges to toxicity evaluation. In this study, a novel strategy was developed to predict the drug-ADR associations by combining deep learning and the biomedical tripartite network. This heterogeneous network contains biomedical linked data of three entities, for example, drugs, targets, and ADRs. For the first time, GraRep, a deep learning method for distributed representations, is introduced to learn graph representations and identify hidden features from the tripartite network which are further used for ADR prediction. Through this approach, drug-ADR associations could possibly be discovered from a systemic perspective. The accuracy of our method is 0.95 based on internal resource validation and 0.88 based on external resource validation. Moreover, our results show the prediction accuracy using the tripartite network is better than the one with bipartite network, suggesting the model performance can be improved with further enrichment on information. According to the result of 10-fold cross validation, the deep learning model outperforms two traditional methods (topology-based measures and chemical structure-based measures). Additionally, predictive models are also constructed using other deep learning methods, and comparable results are achieved. In summary, the biomedical tripartite network-based deep learning model proposed here proves to offer a promising solution for prediction of ADRs.
中文翻译:

结合生物医学三方网络和图表示模型预测药物不良反应。
作为药物临床高流失率的主要贡献者之一,毒性评估对新药的发现至关重要。毫不奇怪,已经在开发流程的各个阶段开发了大量计算方法,以评估潜在的药物不良反应(ADR)。尽管这些方法先前已在单个ADR或某些药物家族中获得成功,但毒性评估仍面临巨大挑战。在这项研究中,通过结合深度学习和生物医学三方网络,开发了一种预测药物-ADR关联的新策略。这个异构网络包含三个实体的生物医学链接数据,例如,药物,靶标和ADR。第一次,GraRep,一种用于分布式表示的深度学习方法,引入来学习图形表示并从三方网络中识别隐藏特征,这些特征将进一步用于ADR预测。通过这种方法,有可能从系统角度发现药物ADR关联。基于内部资源验证,我们方法的准确性为0.95;基于外部资源验证,我们方法的准确性为0.88。此外,我们的结果表明,使用三方网络的预测精度要优于使用二部网络的预测精度,这表明可以通过进一步丰富信息来提高模型性能。根据10倍交叉验证的结果,深度学习模型优于两种传统方法(基于拓扑的度量和基于化学结构的度量)。此外,还可以使用其他深度学习方法来构建预测模型,并获得了可比的结果。总之,这里提出的基于生物医学三方网络的深度学习模型被证明为ADR的预测提供了有希望的解决方案。
更新日期:2019-12-17
中文翻译:

结合生物医学三方网络和图表示模型预测药物不良反应。
作为药物临床高流失率的主要贡献者之一,毒性评估对新药的发现至关重要。毫不奇怪,已经在开发流程的各个阶段开发了大量计算方法,以评估潜在的药物不良反应(ADR)。尽管这些方法先前已在单个ADR或某些药物家族中获得成功,但毒性评估仍面临巨大挑战。在这项研究中,通过结合深度学习和生物医学三方网络,开发了一种预测药物-ADR关联的新策略。这个异构网络包含三个实体的生物医学链接数据,例如,药物,靶标和ADR。第一次,GraRep,一种用于分布式表示的深度学习方法,引入来学习图形表示并从三方网络中识别隐藏特征,这些特征将进一步用于ADR预测。通过这种方法,有可能从系统角度发现药物ADR关联。基于内部资源验证,我们方法的准确性为0.95;基于外部资源验证,我们方法的准确性为0.88。此外,我们的结果表明,使用三方网络的预测精度要优于使用二部网络的预测精度,这表明可以通过进一步丰富信息来提高模型性能。根据10倍交叉验证的结果,深度学习模型优于两种传统方法(基于拓扑的度量和基于化学结构的度量)。此外,还可以使用其他深度学习方法来构建预测模型,并获得了可比的结果。总之,这里提出的基于生物医学三方网络的深度学习模型被证明为ADR的预测提供了有希望的解决方案。











































 京公网安备 11010802027423号
京公网安备 11010802027423号