当前位置:
X-MOL 学术
›
Nat. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Analysis of the Human Protein Atlas Image Classification competition.
Nature Methods ( IF 36.1 ) Pub Date : 2019-11-28 , DOI: 10.1038/s41592-019-0658-6 Wei Ouyang 1 , Casper F Winsnes 1 , Martin Hjelmare 1 , Anthony J Cesnik 2, 3 , Lovisa Åkesson 1 , Hao Xu 1 , Devin P Sullivan 1 , Shubin Dai 4 , Jun Lan 5 , Park Jinmo 6 , Shaikat M Galib 7 , Christof Henkel 8 , Kevin Hwang 9 , Dmytro Poplavskiy 10 , Bojan Tunguz 11 , Russel D Wolfinger 12 , Yinzheng Gu 13 , Chuanpeng Li 13 , Jinbin Xie 13 , Dmitry Buslov 14 , Sergei Fironov 15 , Alexander Kiselev 16 , Dmytro Panchenko 17 , Xuan Cao 18 , Runmin Wei 19 , Yuanhao Wu 20 , Xun Zhu 21 , Kuan-Lun Tseng 22 , Zhifeng Gao 23 , Cheng Ju 24 , Xiaohan Yi 25 , Hongdong Zheng 26 , Constantin Kappel 27 , Emma Lundberg 1, 2, 3
Nature Methods ( IF 36.1 ) Pub Date : 2019-11-28 , DOI: 10.1038/s41592-019-0658-6 Wei Ouyang 1 , Casper F Winsnes 1 , Martin Hjelmare 1 , Anthony J Cesnik 2, 3 , Lovisa Åkesson 1 , Hao Xu 1 , Devin P Sullivan 1 , Shubin Dai 4 , Jun Lan 5 , Park Jinmo 6 , Shaikat M Galib 7 , Christof Henkel 8 , Kevin Hwang 9 , Dmytro Poplavskiy 10 , Bojan Tunguz 11 , Russel D Wolfinger 12 , Yinzheng Gu 13 , Chuanpeng Li 13 , Jinbin Xie 13 , Dmitry Buslov 14 , Sergei Fironov 15 , Alexander Kiselev 16 , Dmytro Panchenko 17 , Xuan Cao 18 , Runmin Wei 19 , Yuanhao Wu 20 , Xun Zhu 21 , Kuan-Lun Tseng 22 , Zhifeng Gao 23 , Cheng Ju 24 , Xiaohan Yi 25 , Hongdong Zheng 26 , Constantin Kappel 27 , Emma Lundberg 1, 2, 3
Affiliation
|
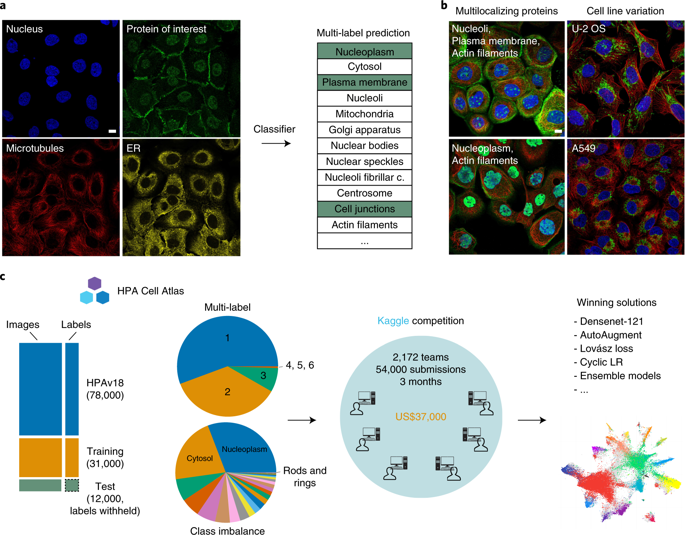
Pinpointing subcellular protein localizations from microscopy images is easy to the trained eye, but challenging to automate. Based on the Human Protein Atlas image collection, we held a competition to identify deep learning solutions to solve this task. Challenges included training on highly imbalanced classes and predicting multiple labels per image. Over 3 months, 2,172 teams participated. Despite convergence on popular networks and training techniques, there was considerable variety among the solutions. Participants applied strategies for modifying neural networks and loss functions, augmenting data and using pretrained networks. The winning models far outperformed our previous effort at multi-label classification of protein localization patterns by ~20%. These models can be used as classifiers to annotate new images, feature extractors to measure pattern similarity or pretrained networks for a wide range of biological applications.
中文翻译:

人类蛋白质图谱图像分类竞赛分析。
对于训练有素的人来说,从显微镜图像中精确定位亚细胞蛋白定位很容易,但自动化却很困难。基于人类蛋白质图谱图像集,我们举办了一场竞赛,以确定解决此任务的深度学习解决方案。挑战包括对高度不平衡的类别进行训练以及预测每个图像的多个标签。超过 3 个月,共有 2,172 支队伍参加。尽管流行网络和训练技术趋同,但解决方案之间存在很大差异。参与者应用了修改神经网络和损失函数、增强数据和使用预训练网络的策略。获胜模型的性能远远超出我们之前在蛋白质定位模式多标签分类方面的成果约 20%。这些模型可用作分类器来注释新图像、特征提取器来测量模式相似性或预训练网络以用于广泛的生物应用。
更新日期:2019-11-29
中文翻译:

人类蛋白质图谱图像分类竞赛分析。
对于训练有素的人来说,从显微镜图像中精确定位亚细胞蛋白定位很容易,但自动化却很困难。基于人类蛋白质图谱图像集,我们举办了一场竞赛,以确定解决此任务的深度学习解决方案。挑战包括对高度不平衡的类别进行训练以及预测每个图像的多个标签。超过 3 个月,共有 2,172 支队伍参加。尽管流行网络和训练技术趋同,但解决方案之间存在很大差异。参与者应用了修改神经网络和损失函数、增强数据和使用预训练网络的策略。获胜模型的性能远远超出我们之前在蛋白质定位模式多标签分类方面的成果约 20%。这些模型可用作分类器来注释新图像、特征提取器来测量模式相似性或预训练网络以用于广泛的生物应用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号