当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Discovering the Microbial Enzymes Driving Drug Toxicity with Activity-Based Protein Profiling.
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2019-12-12 , DOI: 10.1021/acschembio.9b00788 Parth B Jariwala , Samuel J Pellock , Dennis Goldfarb , Erica W Cloer , Marta Artola 1 , Joshua B Simpson , Aadra P Bhatt , William G Walton , Lee R Roberts 2 , Michael B Major , Gideon J Davies 3 , Herman S Overkleeft 1 , Matthew R Redinbo
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2019-12-12 , DOI: 10.1021/acschembio.9b00788 Parth B Jariwala , Samuel J Pellock , Dennis Goldfarb , Erica W Cloer , Marta Artola 1 , Joshua B Simpson , Aadra P Bhatt , William G Walton , Lee R Roberts 2 , Michael B Major , Gideon J Davies 3 , Herman S Overkleeft 1 , Matthew R Redinbo
Affiliation
|
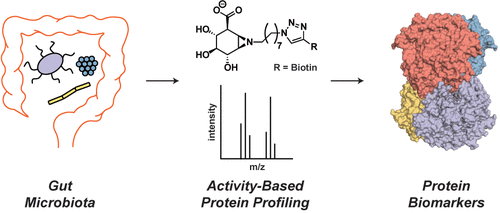
It is increasingly clear that interindividual variability in human gut microbial composition contributes to differential drug responses. For example, gastrointestinal (GI) toxicity is not observed in all patients treated with the anticancer drug irinotecan, and it has been suggested that this variability is a result of differences in the types and levels of gut bacterial β-glucuronidases (GUSs). GUS enzymes promote drug toxicity by hydrolyzing the inactive drug-glucuronide conjugate back to the active drug, which damages the GI epithelium. Proteomics-based identification of the exact GUS enzymes responsible for drug reactivation from the complexity of the human microbiota has not been accomplished, however. Here, we discover the specific bacterial GUS enzymes that generate SN-38, the active and toxic metabolite of irinotecan, from human fecal samples using a unique activity-based protein profiling (ABPP) platform. We identify and quantify gut bacterial GUS enzymes from human feces with an ABPP-enabled proteomics pipeline and then integrate this information with ex vivo kinetics to pinpoint the specific GUS enzymes responsible for SN-38 reactivation. Furthermore, the same approach also reveals the molecular basis for differential gut bacterial GUS inhibition observed between human fecal samples. Taken together, this work provides an unprecedented technical and bioinformatics pipeline to discover the microbial enzymes responsible for specific reactions from the complexity of human feces. Identifying such microbial enzymes may lead to precision biomarkers and novel drug targets to advance the promise of personalized medicine.
中文翻译:

通过基于活性的蛋白质分析发现驱动药物毒性的微生物酶。
越来越清楚的是,人类肠道微生物组成的个体差异导致了不同的药物反应。例如,并非所有接受抗癌药物伊立替康治疗的患者都观察到胃肠道(GI)毒性,有人认为这种变异性是肠道细菌β-葡萄糖醛酸酶(GUS)类型和水平差异的结果。 GUS 酶通过将非活性药物-葡萄糖醛酸结合物水解回活性药物来促进药物毒性,从而损害胃肠道上皮。然而,基于蛋白质组学的人类微生物群复杂性中负责药物再激活的确切 GUS 酶的鉴定尚未完成。在这里,我们使用独特的基于活性的蛋白质分析 (ABPP) 平台从人类粪便样本中发现了产生 SN-38(伊立替康的活性和有毒代谢物)的特定细菌 GUS 酶。我们通过支持 ABPP 的蛋白质组学流程识别和量化人类粪便中的肠道细菌 GUS 酶,然后将这些信息与离体动力学相结合,以查明负责 SN-38 重新激活的特定 GUS 酶。此外,同样的方法还揭示了在人类粪便样本中观察到的差异肠道细菌 GUS 抑制的分子基础。总而言之,这项工作提供了前所未有的技术和生物信息学途径,以发现负责人类粪便复杂性中特定反应的微生物酶。识别此类微生物酶可能会产生精确的生物标志物和新的药物靶点,从而推进个性化医疗的前景。
更新日期:2019-12-13
中文翻译:

通过基于活性的蛋白质分析发现驱动药物毒性的微生物酶。
越来越清楚的是,人类肠道微生物组成的个体差异导致了不同的药物反应。例如,并非所有接受抗癌药物伊立替康治疗的患者都观察到胃肠道(GI)毒性,有人认为这种变异性是肠道细菌β-葡萄糖醛酸酶(GUS)类型和水平差异的结果。 GUS 酶通过将非活性药物-葡萄糖醛酸结合物水解回活性药物来促进药物毒性,从而损害胃肠道上皮。然而,基于蛋白质组学的人类微生物群复杂性中负责药物再激活的确切 GUS 酶的鉴定尚未完成。在这里,我们使用独特的基于活性的蛋白质分析 (ABPP) 平台从人类粪便样本中发现了产生 SN-38(伊立替康的活性和有毒代谢物)的特定细菌 GUS 酶。我们通过支持 ABPP 的蛋白质组学流程识别和量化人类粪便中的肠道细菌 GUS 酶,然后将这些信息与离体动力学相结合,以查明负责 SN-38 重新激活的特定 GUS 酶。此外,同样的方法还揭示了在人类粪便样本中观察到的差异肠道细菌 GUS 抑制的分子基础。总而言之,这项工作提供了前所未有的技术和生物信息学途径,以发现负责人类粪便复杂性中特定反应的微生物酶。识别此类微生物酶可能会产生精确的生物标志物和新的药物靶点,从而推进个性化医疗的前景。











































 京公网安备 11010802027423号
京公网安备 11010802027423号