Cell Systems ( IF 9.0 ) Pub Date : 2019-11-27 , DOI: 10.1016/j.cels.2019.10.009 Sam F Greenbury 1 , Mauricio Barahona 2 , Iain G Johnston 3
|
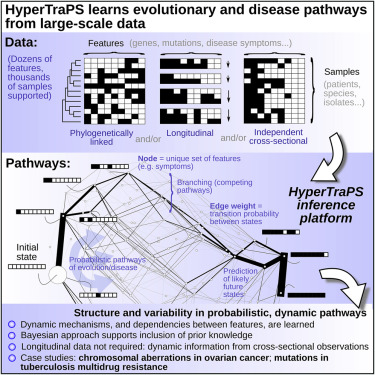
The explosion of data throughout the biomedical sciences provides unprecedented opportunities to learn about the dynamics of evolution and disease progression, but harnessing these large and diverse datasets remains challenging. Here, we describe a highly generalizable statistical platform to infer the dynamic pathways by which many, potentially interacting, traits are acquired or lost over time. We use HyperTraPS (hypercubic transition path sampling) to efficiently learn progression pathways from cross-sectional, longitudinal, or phylogenetically linked data, readily distinguishing multiple competing pathways, and identifying the most parsimonious mechanisms underlying given observations. This Bayesian approach allows inclusion of prior knowledge, quantifies uncertainty in pathway structure, and allows predictions, such as which symptom a patient will acquire next. We provide visualization tools for intuitive assessment of multiple, variable pathways. We apply the method to ovarian cancer progression and the evolution of multidrug resistance in tuberculosis, demonstrating its power to reveal previously undetected dynamic pathways.
中文翻译:

HyperTraPS:在进化和疾病进展途径中推断性状习性的概率模式。
整个生物医学科学领域的数据爆炸提供了前所未有的机会来了解进化和疾病进展的动态,但是如何利用这些庞大而多样的数据集仍然具有挑战性。在这里,我们描述了一个高度可概括的统计平台,以推断随着时间的推移获取或丢失许多可能交互作用的性状的动态途径。我们使用HyperTraPS(高三次过渡路径采样)从横截面,纵向或系统发育关联的数据中有效地了解进展途径,轻松地区分多种竞争途径,并确定给定观察结果的最简约机制。这种贝叶斯方法允许包含先验知识,量化路径结构中的不确定性,并可以进行预测,例如患者接下来会出现哪种症状。我们提供可视化工具,用于直观评估多个可变路径。我们将该方法应用于卵巢癌的进展和结核病多药耐药性的演变,证明了其揭示以前未发现的动态途径的能力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号