当前位置:
X-MOL 学术
›
Nat. Protoc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP).
Nature Protocols ( IF 14.8 ) Pub Date : 2019-11-20 , DOI: 10.1038/s41596-019-0227-6 Yichi Zhang 1 , Tianrun Cai 2 , Sheng Yu 3, 4 , Kelly Cho 5, 6 , Chuan Hong 1 , Jiehuan Sun 1 , Jie Huang 2 , Yuk-Lam Ho 5 , Ashwin N Ananthakrishnan 7 , Zongqi Xia 8 , Stanley Y Shaw 9 , Vivian Gainer 10 , Victor Castro 10 , Nicholas Link 5 , Jacqueline Honerlaw 5 , Sicong Huang 2 , David Gagnon 5, 11 , Elizabeth W Karlson 2 , Robert M Plenge 2 , Peter Szolovits 12 , Guergana Savova 13 , Susanne Churchill 14 , Christopher O'Donnell 5, 15 , Shawn N Murphy 10, 14, 16 , J Michael Gaziano 5, 6 , Isaac Kohane 14 , Tianxi Cai 1, 14 , Katherine P Liao 2, 5, 14
Nature Protocols ( IF 14.8 ) Pub Date : 2019-11-20 , DOI: 10.1038/s41596-019-0227-6 Yichi Zhang 1 , Tianrun Cai 2 , Sheng Yu 3, 4 , Kelly Cho 5, 6 , Chuan Hong 1 , Jiehuan Sun 1 , Jie Huang 2 , Yuk-Lam Ho 5 , Ashwin N Ananthakrishnan 7 , Zongqi Xia 8 , Stanley Y Shaw 9 , Vivian Gainer 10 , Victor Castro 10 , Nicholas Link 5 , Jacqueline Honerlaw 5 , Sicong Huang 2 , David Gagnon 5, 11 , Elizabeth W Karlson 2 , Robert M Plenge 2 , Peter Szolovits 12 , Guergana Savova 13 , Susanne Churchill 14 , Christopher O'Donnell 5, 15 , Shawn N Murphy 10, 14, 16 , J Michael Gaziano 5, 6 , Isaac Kohane 14 , Tianxi Cai 1, 14 , Katherine P Liao 2, 5, 14
Affiliation
|
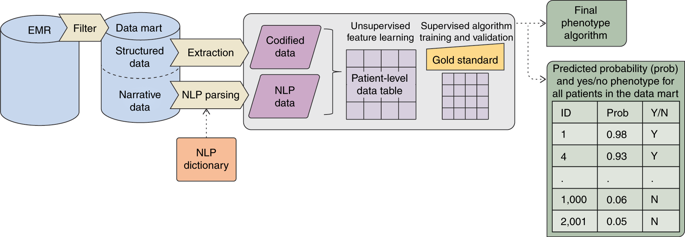
Phenotypes are the foundation for clinical and genetic studies of disease risk and outcomes. The growth of biobanks linked to electronic medical record (EMR) data has both facilitated and increased the demand for efficient, accurate, and robust approaches for phenotyping millions of patients. Challenges to phenotyping with EMR data include variation in the accuracy of codes, as well as the high level of manual input required to identify features for the algorithm and to obtain gold standard labels. To address these challenges, we developed PheCAP, a high-throughput semi-supervised phenotyping pipeline. PheCAP begins with data from the EMR, including structured data and information extracted from the narrative notes using natural language processing (NLP). The standardized steps integrate automated procedures, which reduce the level of manual input, and machine learning approaches for algorithm training. PheCAP itself can be executed in 1-2 d if all data are available; however, the timing is largely dependent on the chart review stage, which typically requires at least 2 weeks. The final products of PheCAP include a phenotype algorithm, the probability of the phenotype for all patients, and a phenotype classification (yes or no).
中文翻译:

使用常见的半监督方法 (PheCAP) 对电子病历数据进行高通量表型分析。
表型是疾病风险和结果的临床和遗传学研究的基础。与电子病历 (EMR) 数据相关的生物库的增长促进并增加了对数百万患者表型分析的高效、准确和稳健方法的需求。使用 EMR 数据进行表型分析的挑战包括代码准确性的变化,以及识别算法特征和获得金标准标签所需的高水平手动输入。为了应对这些挑战,我们开发了 PheCAP,一种高通量半监督表型分析管道。PheCAP 从 EMR 中的数据开始,包括结构化数据和使用自然语言处理 (NLP) 从叙述笔记中提取的信息。标准化的步骤集成了自动化程序,从而降低了手动输入的水平,和用于算法训练的机器学习方法。如果所有数据都可用,PheCAP 本身可以在 1-2 d 内执行;但是,时间主要取决于图表审查阶段,通常需要至少 2 周。PheCAP 的最终产品包括表型算法、所有患者的表型概率和表型分类(是或否)。
更新日期:2019-11-21
中文翻译:

使用常见的半监督方法 (PheCAP) 对电子病历数据进行高通量表型分析。
表型是疾病风险和结果的临床和遗传学研究的基础。与电子病历 (EMR) 数据相关的生物库的增长促进并增加了对数百万患者表型分析的高效、准确和稳健方法的需求。使用 EMR 数据进行表型分析的挑战包括代码准确性的变化,以及识别算法特征和获得金标准标签所需的高水平手动输入。为了应对这些挑战,我们开发了 PheCAP,一种高通量半监督表型分析管道。PheCAP 从 EMR 中的数据开始,包括结构化数据和使用自然语言处理 (NLP) 从叙述笔记中提取的信息。标准化的步骤集成了自动化程序,从而降低了手动输入的水平,和用于算法训练的机器学习方法。如果所有数据都可用,PheCAP 本身可以在 1-2 d 内执行;但是,时间主要取决于图表审查阶段,通常需要至少 2 周。PheCAP 的最终产品包括表型算法、所有患者的表型概率和表型分类(是或否)。



























 京公网安备 11010802027423号
京公网安备 11010802027423号