Cell Systems ( IF 9.0 ) Pub Date : 2019-11-13 , DOI: 10.1016/j.cels.2019.10.004 Martin Lukačišin 1 , Tobias Bollenbach 2
|
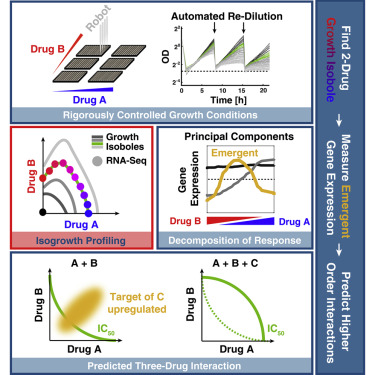
Effective design of combination therapies requires understanding the changes in cell physiology that result from drug interactions. Here, we show that the genome-wide transcriptional response to combinations of two drugs, measured at a rigorously controlled growth rate, can predict higher-order antagonism with a third drug in Saccharomyces cerevisiae. Using isogrowth profiling, over 90% of the variation in cellular response can be decomposed into three principal components (PCs) that have clear biological interpretations. We demonstrate that the third PC captures emergent transcriptional programs that are dependent on both drugs and can predict antagonism with a third drug targeting the emergent pathway. We further show that emergent gene expression patterns are most pronounced at a drug ratio where the drug interaction is strongest, providing a guideline for future measurements. Our results provide a readily applicable recipe for uncovering emergent responses in other systems and for higher-order drug combinations. A record of this paper’s transparent peer review process is included in the Supplemental Information.
中文翻译:

对药物组合的新兴基因表达反应可预测更高阶的药物相互作用。
有效的联合疗法设计需要了解药物相互作用引起的细胞生理学变化。在这里,我们表明,在严格控制的生长速率下,对两种药物组合的全基因组转录反应可以预测酿酒酵母中与第三种药物的更高程度的拮抗作用。。使用等长线图分析,细胞反应中超过90%的变化可以分解为具有清晰生物学解释的三个主要成分(PC)。我们证明,第三台PC捕获依赖于两种药物的紧急转录程序,并且可以预测与靶向紧急途径的第三种药物的拮抗作用。我们进一步表明,在药物相互作用最强的药物比例下,出现的基因表达模式最为明显,为将来的测量提供了指导。我们的结果为发现其他系统中的紧急反应和更高级别的药物组合提供了易于应用的方法。补充信息中包含了本文透明的同行评审过程的记录。











































 京公网安备 11010802027423号
京公网安备 11010802027423号