Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Modeling Antibody-Antigen Complexes by Information-Driven Docking.
Structure ( IF 4.4 ) Pub Date : 2019-11-11 , DOI: 10.1016/j.str.2019.10.011 Francesco Ambrosetti 1 , Brian Jiménez-García 2 , Jorge Roel-Touris 2 , Alexandre M J J Bonvin 2
Structure ( IF 4.4 ) Pub Date : 2019-11-11 , DOI: 10.1016/j.str.2019.10.011 Francesco Ambrosetti 1 , Brian Jiménez-García 2 , Jorge Roel-Touris 2 , Alexandre M J J Bonvin 2
Affiliation
|
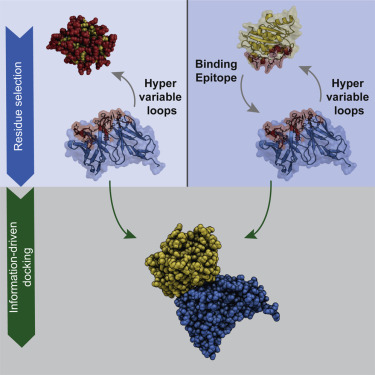
Antibodies are Y-shaped proteins essential for immune response. Their capability to recognize antigens with high specificity makes them excellent therapeutic targets. Understanding the structural basis of antibody-antigen interactions is therefore crucial for improving our ability to design efficient biological drugs. Computational approaches such as molecular docking are providing a valuable and fast alternative to experimental structural characterization for these complexes. We investigate here how information about complementarity-determining regions and binding epitopes can be used to drive the modeling process, and present a comparative study of four different docking software suites (ClusPro, LightDock, ZDOCK, and HADDOCK) providing specific options for antibody-antigen modeling. Their performance on a dataset of 16 complexes is reported. HADDOCK, which includes information to drive the docking, is shown to perform best in terms of both success rate and quality of the generated models in both the presence and absence of information about the epitope on the antigen.
中文翻译:

通过信息驱动对接对抗体-抗原复合物进行建模。
抗体是免疫反应必不可少的Y型蛋白。它们以高特异性识别抗原的能力使其成为极好的治疗靶标。因此,了解抗体-抗原相互作用的结构基础对于提高我们设计有效生物药物的能力至关重要。诸如分子对接之类的计算方法为这些复合物的实验结构表征提供了一种有价值且快速的替代方法。我们在这里研究如何将有关互补决定区和结合表位的信息用于驱动建模过程,并对四种不同的对接软件套件(ClusPro,LightDock,ZDOCK和HADDOCK)进行比较研究,为抗体-抗原提供了特定的选择造型。报告了它们在16种配合物的数据集上的性能。
更新日期:2019-11-13
中文翻译:

通过信息驱动对接对抗体-抗原复合物进行建模。
抗体是免疫反应必不可少的Y型蛋白。它们以高特异性识别抗原的能力使其成为极好的治疗靶标。因此,了解抗体-抗原相互作用的结构基础对于提高我们设计有效生物药物的能力至关重要。诸如分子对接之类的计算方法为这些复合物的实验结构表征提供了一种有价值且快速的替代方法。我们在这里研究如何将有关互补决定区和结合表位的信息用于驱动建模过程,并对四种不同的对接软件套件(ClusPro,LightDock,ZDOCK和HADDOCK)进行比较研究,为抗体-抗原提供了特定的选择造型。报告了它们在16种配合物的数据集上的性能。











































 京公网安备 11010802027423号
京公网安备 11010802027423号