当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
From expression footprints to causal pathways: contextualizing large signaling networks with CARNIVAL.
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-11-11 , DOI: 10.1038/s41540-019-0118-z Anika Liu 1, 2 , Panuwat Trairatphisan 1 , Enio Gjerga 1, 2 , Athanasios Didangelos 3 , Jonathan Barratt 3 , Julio Saez-Rodriguez 1, 2
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-11-11 , DOI: 10.1038/s41540-019-0118-z Anika Liu 1, 2 , Panuwat Trairatphisan 1 , Enio Gjerga 1, 2 , Athanasios Didangelos 3 , Jonathan Barratt 3 , Julio Saez-Rodriguez 1, 2
Affiliation
|
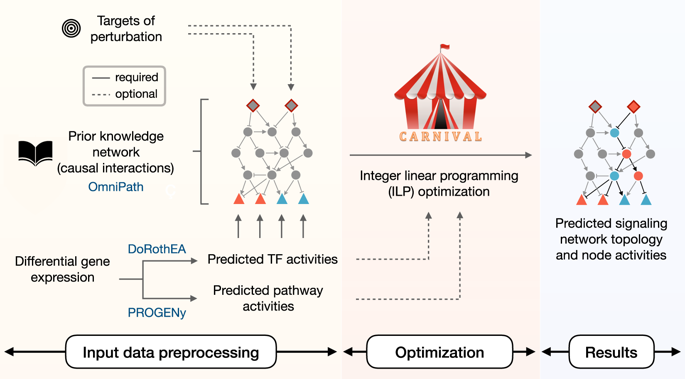
While gene expression profiling is commonly used to gain an overview of cellular processes, the identification of upstream processes that drive expression changes remains a challenge. To address this issue, we introduce CARNIVAL, a causal network contextualization tool which derives network architectures from gene expression footprints. CARNIVAL (CAusal Reasoning pipeline for Network identification using Integer VALue programming) integrates different sources of prior knowledge including signed and directed protein-protein interactions, transcription factor targets, and pathway signatures. The use of prior knowledge in CARNIVAL enables capturing a broad set of upstream cellular processes and regulators, leading to a higher accuracy when benchmarked against related tools. Implementation as an integer linear programming (ILP) problem guarantees efficient computation. As a case study, we applied CARNIVAL to contextualize signaling networks from gene expression data in IgA nephropathy (IgAN), a condition that can lead to chronic kidney disease. CARNIVAL identified specific signaling pathways and associated mediators dysregulated in IgAN including Wnt and TGF-β, which we subsequently validated experimentally. These results demonstrated how CARNIVAL generates hypotheses on potential upstream alterations that propagate through signaling networks, providing insights into diseases.
中文翻译:

从表达足迹到因果路径:将大型信号网络与嘉年华结合起来。
虽然基因表达谱通常用于获得细胞过程的概述,但驱动表达变化的上游过程的识别仍然是一个挑战。为了解决这个问题,我们引入了 CARNIVAL,一种因果网络情境化工具,可以从基因表达足迹中导出网络架构。 CARNIVAL(使用整数值编程进行网络识别的 CAusal 推理管道)集成了不同来源的先验知识,包括有符号和定向的蛋白质-蛋白质相互作用、转录因子目标和途径签名。在 CARNIVAL 中使用先验知识可以捕获广泛的上游细胞过程和调节器,从而在与相关工具进行基准测试时获得更高的准确性。作为整数线性规划 (ILP) 问题的实现保证了高效计算。作为一个案例研究,我们应用 CARNIVAL 来了解 IgA 肾病 (IgAN) 基因表达数据中的信号网络,IgAN 是一种可能导致慢性肾脏疾病的疾病。 CARNIVAL 确定了 IgAN 中失调的特定信号通路和相关介质,包括 Wnt 和 TGF-β,我们随后进行了实验验证。这些结果证明了 CARNIVAL 如何对通过信号网络传播的潜在上游变化产生假设,从而提供对疾病的见解。
更新日期:2019-11-11
中文翻译:

从表达足迹到因果路径:将大型信号网络与嘉年华结合起来。
虽然基因表达谱通常用于获得细胞过程的概述,但驱动表达变化的上游过程的识别仍然是一个挑战。为了解决这个问题,我们引入了 CARNIVAL,一种因果网络情境化工具,可以从基因表达足迹中导出网络架构。 CARNIVAL(使用整数值编程进行网络识别的 CAusal 推理管道)集成了不同来源的先验知识,包括有符号和定向的蛋白质-蛋白质相互作用、转录因子目标和途径签名。在 CARNIVAL 中使用先验知识可以捕获广泛的上游细胞过程和调节器,从而在与相关工具进行基准测试时获得更高的准确性。作为整数线性规划 (ILP) 问题的实现保证了高效计算。作为一个案例研究,我们应用 CARNIVAL 来了解 IgA 肾病 (IgAN) 基因表达数据中的信号网络,IgAN 是一种可能导致慢性肾脏疾病的疾病。 CARNIVAL 确定了 IgAN 中失调的特定信号通路和相关介质,包括 Wnt 和 TGF-β,我们随后进行了实验验证。这些结果证明了 CARNIVAL 如何对通过信号网络传播的潜在上游变化产生假设,从而提供对疾病的见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号