当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Computationally Guided Discovery and Experimental Validation of Indole-3-acetic Acid Synthesis Pathways.
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2019-11-18 , DOI: 10.1021/acschembio.9b00725 David C Garcia 1, 2 , Xiaolin Cheng 3 , Miriam L Land 4 , Robert F Standaert 1, 5 , Jennifer L Morrell-Falvey 1 , Mitchel J Doktycz 1, 2
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2019-11-18 , DOI: 10.1021/acschembio.9b00725 David C Garcia 1, 2 , Xiaolin Cheng 3 , Miriam L Land 4 , Robert F Standaert 1, 5 , Jennifer L Morrell-Falvey 1 , Mitchel J Doktycz 1, 2
Affiliation
|
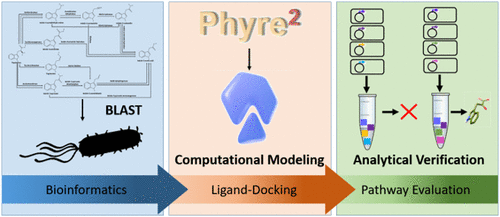
Elucidating the interaction networks associated with secondary metabolite production in microorganisms is an ongoing challenge made all the more daunting by the rate at which DNA sequencing technology reveals new genes and potential pathways. Developing the culturing methods, expression conditions, and genetic systems needed for validating pathways in newly discovered microorganisms is often not possible. Therefore, new tools and techniques are needed for defining complex metabolic pathways. Here, we describe an in vitro computationally assisted pathway description approach that employs bioinformatic searches of genome databases, protein structural modeling, and protein-ligand-docking simulations to predict the gene products most likely to be involved in a particular secondary metabolite production pathway. This information is then used to direct in vitro reconstructions of the pathway and subsequent confirmation of pathway activity using crude enzyme preparations. As a test system, we elucidated the pathway for biosynthesis of indole-3-acetic acid (IAA) in the plant-associated microbe Pantoea sp. YR343. This organism is capable of metabolizing tryptophan into the plant phytohormone IAA. BLAST analyses identified a likely three-step pathway involving an amino transferase, an indole pyruvate decarboxylase, and a dehydrogenase. However, multiple candidate enzymes were identified at each step, resulting in a large number of potential pathway reconstructions (32 different enzyme combinations). Our approach shows the effectiveness of crude extracts to rapidly elucidate enzymes leading to functional pathways. Results are compared to affinity purified enzymes for select combinations and found to yield similar relative activities. Further, in vitro testing of the pathway reconstructions revealed the "underground" nature of IAA metabolism in Pantoea sp. YR343 and the various mechanisms used to produce IAA. Importantly, our experiments illustrate the scalable integration of computational tools and cell-free enzymatic reactions to identify and validate metabolic pathways in a broadly applicable manner.
中文翻译:

吲哚-3-乙酸合成途径的计算指导发现和实验验证。
阐明与微生物中次级代谢产物产生相关的相互作用网络是一项持续的挑战,而DNA测序技术揭示新基因和潜在途径的速度使这一挑战变得更加艰巨。通常不可能开发验证新发现的微生物中的途径所需的培养方法,表达条件和遗传系统。因此,需要新的工具和技术来定义复杂的代谢途径。在这里,我们描述了一种体外计算机辅助途径描述方法,该方法利用基因组数据库的生物信息学搜索,蛋白质结构建模和蛋白质配体对接模拟来预测最有可能参与特定次级代谢产物生产途径的基因产物。然后,该信息将用于指导该途径的体外重建,并随后使用粗酶制剂确认该途径的活性。作为测试系统,我们阐明了植物相关微生物Pantoea sp。中吲哚-3-乙酸(IAA)生物合成的途径。YR343。该生物能够将色氨酸代谢为植物植物激素IAA。BLAST分析确定了可能的三步途径,涉及氨基转移酶,吲哚丙酮酸脱羧酶和脱氢酶。然而,在每个步骤中都鉴定出多种候选酶,从而导致大量潜在的途径重建(32种不同的酶组合)。我们的方法显示了粗提物快速阐明导致功能途径的酶的有效性。将结果与亲和纯化的酶进行选择组合进行比较,发现产生相似的相对活性。此外,对途径重建的体外测试揭示了Pantoea sp。中IAA代谢的“地下”性质。YR343和用于产生IAA的各种机制。重要的是,我们的实验说明了计算工具和无细胞酶促反应的可扩展集成,以广泛适用的方式识别和验证代谢途径。
更新日期:2019-11-18
中文翻译:

吲哚-3-乙酸合成途径的计算指导发现和实验验证。
阐明与微生物中次级代谢产物产生相关的相互作用网络是一项持续的挑战,而DNA测序技术揭示新基因和潜在途径的速度使这一挑战变得更加艰巨。通常不可能开发验证新发现的微生物中的途径所需的培养方法,表达条件和遗传系统。因此,需要新的工具和技术来定义复杂的代谢途径。在这里,我们描述了一种体外计算机辅助途径描述方法,该方法利用基因组数据库的生物信息学搜索,蛋白质结构建模和蛋白质配体对接模拟来预测最有可能参与特定次级代谢产物生产途径的基因产物。然后,该信息将用于指导该途径的体外重建,并随后使用粗酶制剂确认该途径的活性。作为测试系统,我们阐明了植物相关微生物Pantoea sp。中吲哚-3-乙酸(IAA)生物合成的途径。YR343。该生物能够将色氨酸代谢为植物植物激素IAA。BLAST分析确定了可能的三步途径,涉及氨基转移酶,吲哚丙酮酸脱羧酶和脱氢酶。然而,在每个步骤中都鉴定出多种候选酶,从而导致大量潜在的途径重建(32种不同的酶组合)。我们的方法显示了粗提物快速阐明导致功能途径的酶的有效性。将结果与亲和纯化的酶进行选择组合进行比较,发现产生相似的相对活性。此外,对途径重建的体外测试揭示了Pantoea sp。中IAA代谢的“地下”性质。YR343和用于产生IAA的各种机制。重要的是,我们的实验说明了计算工具和无细胞酶促反应的可扩展集成,以广泛适用的方式识别和验证代谢途径。











































 京公网安备 11010802027423号
京公网安备 11010802027423号