当前位置:
X-MOL 学术
›
Mol. Phylogenet. Evol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Cross-contamination and strong mitonuclear discordance in Empria sawflies (Hymenoptera, Tenthredinidae) in the light of phylogenomic data.
Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2019-11-06 , DOI: 10.1016/j.ympev.2019.106670 Marko Prous 1 , Kyung Min Lee 2 , Marko Mutanen 2
Molecular Phylogenetics and Evolution ( IF 4.1 ) Pub Date : 2019-11-06 , DOI: 10.1016/j.ympev.2019.106670 Marko Prous 1 , Kyung Min Lee 2 , Marko Mutanen 2
Affiliation
|
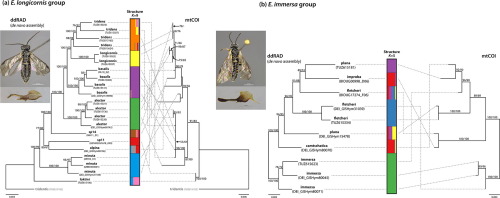
In several sawfly taxa strong mitonuclear discordance has been observed, with nuclear genes supporting species assignments based on morphology, whereas the barcode region of the mitochondrial COI gene suggests different relationships. As previous studies were based on only a few nuclear genes, the causes and the degree of mitonuclear discordance remain ambiguous. Here, we obtained genomic-scale ddRAD data together with Sanger sequences of mitochondrial COI and two to three nuclear protein coding genes to investigate species limits and mitonuclear discordance in two closely related species groups of the sawfly genus Empria. As found previously based on nuclear ITS and mitochondrial COI sequences, species are in most cases supported as monophyletic based on new nuclear data reported here, but not based on mitochondrial COI. This mitonuclear discordance can be explained by occasional mitochondrial introgression with little or no nuclear gene flow, a pattern that might be common in haplodiploid taxa with slowly evolving mitochondrial genomes. Some species in the E. immersa group are not recovered as monophyletic according to either mitochondrial or nuclear data, but this could partly be because of unresolved taxonomy. Preliminary analyses of ddRAD data did not recover monophyly of E. japonica within the E. longicornis group (three Sanger sequenced nuclear genes strongly supported monophyly), but closer examination of the data and additional Sanger sequencing suggested that both specimens were substantially (possibly 10-20% of recovered loci) cross-contaminated. A reason could be specimen identification tag jumps during sequencing library preparation that in previous studies have been shown to affect up to 2.5% of the sequenced reads. We provide an R script to examine patterns of identical loci among the specimens and estimate that the cross-contamination rate is not unusually high for our ddRAD dataset as a whole (based on counting of identical sequences in the immersa and longicornis groups, which are well separated from each other and probably do not hybridise). The high rate of cross-contamination for both E. japonica specimens might be explained by the small number of recovered loci (~1000) compared to most other specimens (>10 000 in some cases) because of poor sequencing results. We caution against drawing unexpected biological conclusions when closely related specimens are pooled before sequencing and tagged only at one end of the molecule or at both ends using a unique combination of limited number of tags (less than the number of specimens).
中文翻译:

根据植物学数据,在Empria锯蝇(膜翅目,天蛾科)中存在交叉污染和强烈的线粒体不一致。
在几个锯齿类群中,已经观察到强烈的线粒体不一致,其核基因支持基于形态的物种分配,而线粒体COI基因的条形码区域暗示了不同的关系。由于先前的研究仅基于少数核基因,因此微核不和谐的原因和程度仍然不明确。在这里,我们获得了基因组规模的ddRAD数据,以及线粒体COI的Sanger序列和2至3个核蛋白编码基因,以研究锯齿蝇Empria的两个密切相关物种组的物种极限和线粒体不一致。正如先前基于核ITS和线粒体COI序列所发现的那样,在大多数情况下,根据此处报告的新核数据,将物种支持为单系物种,而不是基于线粒体COI。这种线粒体不一致的现象可以通过偶发的线粒体渗入而很少或没有核基因流来解释,这种模式在线粒体基因组缓慢进化的单倍体类群中可能很常见。根据线粒体或核数据,E。immersa组中的某些物种未恢复为单系物种,但这可能部分是由于未解决的分类学。对ddRAD数据的初步分析未发现长假肠埃希菌组中的日本大肠埃希菌(三个桑格测序核基因得到了强烈的单生物学支持),但是仔细检查数据和附加的桑格测序表明,两个标本基本相同(可能为10- 20%的回收基因座被交叉污染。一个原因可能是在测序文库制备过程中标本识别标签跳跃,在先前的研究中已显示影响多达2.5%的测序读段。我们提供了一个R脚本来检查样本中相同基因座的模式,并估计整个ddRAD数据集的交叉污染率并非异常高(基于对immersa和longicornis组中相同序列的计数,彼此分离并且可能不会杂交)。由于测序结果差,与大多数其他标本(某些情况下> 10,000)相比,回收的基因座数量少(〜1000),可以解释这两个粳稻标本的高交叉污染率。
更新日期:2019-11-06
中文翻译:

根据植物学数据,在Empria锯蝇(膜翅目,天蛾科)中存在交叉污染和强烈的线粒体不一致。
在几个锯齿类群中,已经观察到强烈的线粒体不一致,其核基因支持基于形态的物种分配,而线粒体COI基因的条形码区域暗示了不同的关系。由于先前的研究仅基于少数核基因,因此微核不和谐的原因和程度仍然不明确。在这里,我们获得了基因组规模的ddRAD数据,以及线粒体COI的Sanger序列和2至3个核蛋白编码基因,以研究锯齿蝇Empria的两个密切相关物种组的物种极限和线粒体不一致。正如先前基于核ITS和线粒体COI序列所发现的那样,在大多数情况下,根据此处报告的新核数据,将物种支持为单系物种,而不是基于线粒体COI。这种线粒体不一致的现象可以通过偶发的线粒体渗入而很少或没有核基因流来解释,这种模式在线粒体基因组缓慢进化的单倍体类群中可能很常见。根据线粒体或核数据,E。immersa组中的某些物种未恢复为单系物种,但这可能部分是由于未解决的分类学。对ddRAD数据的初步分析未发现长假肠埃希菌组中的日本大肠埃希菌(三个桑格测序核基因得到了强烈的单生物学支持),但是仔细检查数据和附加的桑格测序表明,两个标本基本相同(可能为10- 20%的回收基因座被交叉污染。一个原因可能是在测序文库制备过程中标本识别标签跳跃,在先前的研究中已显示影响多达2.5%的测序读段。我们提供了一个R脚本来检查样本中相同基因座的模式,并估计整个ddRAD数据集的交叉污染率并非异常高(基于对immersa和longicornis组中相同序列的计数,彼此分离并且可能不会杂交)。由于测序结果差,与大多数其他标本(某些情况下> 10,000)相比,回收的基因座数量少(〜1000),可以解释这两个粳稻标本的高交叉污染率。



























 京公网安备 11010802027423号
京公网安备 11010802027423号