Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2019-10-23 , DOI: 10.1016/j.jbi.2019.103318 Cong Liu 1 , Casey N Ta 1 , James R Rogers 1 , Ziran Li 1 , Junghwan Lee 1 , Alex M Butler 1 , Ning Shang 1 , Fabricio Sampaio Peres Kury 1 , Liwei Wang 2 , Feichen Shen 2 , Hongfang Liu 2 , Lyudmila Ena 1 , Carol Friedman 1 , Chunhua Weng 1
|
Background
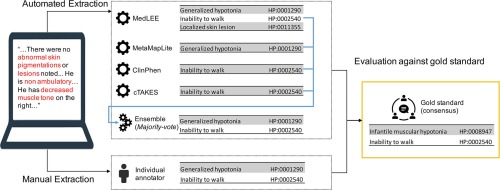
Manually curating standardized phenotypic concepts such as Human Phenotype Ontology (HPO) terms from narrative text in electronic health records (EHRs) is time consuming and error prone. Natural language processing (NLP) techniques can facilitate automated phenotype extraction and thus improve the efficiency of curating clinical phenotypes from clinical texts. While individual NLP systems can perform well for a single cohort, an ensemble-based method might shed light on increasing the portability of NLP pipelines across different cohorts.
Methods
We compared four NLP systems, MetaMapLite, MedLEE, ClinPhen and cTAKES, and four ensemble techniques, including intersection, union, majority-voting and machine learning, for extracting generic phenotypic concepts. We addressed two important research questions regarding automated phenotype recognition. First, we evaluated the performance of different approaches in identifying generic phenotypic concepts. Second, we compared the performance of different methods to identify patient-specific phenotypic concepts. To better quantify the effects caused by concept granularity differences on performance, we developed a novel evaluation metric that considered concept hierarchies and frequencies. Each of the approaches was evaluated on a gold standard set of clinical documents annotated by clinical experts. One dataset containing 1,609 concepts derived from 50 clinical notes from two different institutions was used in both evaluations, and an additional dataset of 608 concepts derived from 50 case report abstracts obtained from PubMed was used for evaluation of identifying generic phenotypic concepts only.
Results
For generic phenotypic concept recognition, the top three performers in the NYP/CUIMC dataset are union ensemble (F1, 0.634), training-based ensemble (F1, 0.632), and majority vote-based ensemble (F1, 0.622). In the Mayo dataset, the top three are majority vote-based ensemble (F1, 0.642), cTAKES (F1, 0.615), and MedLEE (F1, 0.559). In the PubMed dataset, the top three are majority vote-based ensemble (F1, 0.719), training-based (F1, 0.696) and MetaMapLite (F1, 0.694). For identifying patient specific phenotypes, the top three performers in the NYP/CUIMC dataset are majority vote-based ensemble (F1, 0.610), MedLEE (F1, 0.609), and training-based ensemble (F1, 0.585). In the Mayo dataset, the top three are majority vote-based ensemble (F1, 0.604), cTAKES (F1, 0.531) and MedLEE (F1, 0.527).
Conclusions
Our study demonstrates that ensembles of natural language processing can improve both generic phenotypic concept recognition and patient specific phenotypic concept identification over individual systems. Among the individual NLP systems, each individual system performed best when they were applied in the dataset that they were primary designed for. However, combining multiple NLP systems to create an ensemble can generally improve the performance. Specifically, the ensemble can increase the results reproducibility across different cohorts and tasks, and thus provide a more portable phenotyping solution compared to individual NLP systems.
中文翻译:

用于便携式表型解决方案的自然语言处理系统的集合。
背景
从电子健康记录(EHR)的叙述文本中手动制定标准化的表型概念(例如人类表型本体论(HPO)术语)既费时又容易出错。自然语言处理(NLP)技术可以促进自动表型的提取,从而提高从临床文本中整理临床表型的效率。尽管单个NLP系统可以在单个队列中表现良好,但是基于集成的方法可能有助于提高NLP管道在不同队列中的可移植性。
方法
我们比较了四个NLP系统,MetaMapLite,MedLEE,ClinPhen和cTAKES,以及四个系统集成技术,包括交集,并集,多数投票和机器学习,以提取通用表型概念。我们针对自动表型识别解决了两个重要的研究问题。首先,我们评估了识别通用表型概念的不同方法的性能。其次,我们比较了不同方法识别患者特定表型概念的性能。为了更好地量化概念粒度差异对性能造成的影响,我们开发了一种新颖的评估指标,其中考虑了概念层次结构和频率。每种方法均在临床专家注释的临床标准黄金标准集上进行了评估。一个包含1个数据集
结果
对于一般的表型概念的认可,在NYP / CUIMC数据集是工会合奏前三名表演(F 1,0.634),培训为基础的合奏(F 1,0.632),而多数投票为主合奏(F 1,0.622)。在梅奥数据集,前三个是基于表决多数合奏(F 1,0.642),cTAKES(F 1,0.615mmol),和MedLEE(F 1,0.559)。在数据集PUBMED,前三位是多数表决基于合奏(F 1,0.719),训练基于(F 1,0.696mmol)和MetaMapLite(F 1,0.694)。为了识别患者的特定表型,NYP / CUIMC数据集中表现最好的前三名是基于多数投票的合奏(F1,0.610),MedLEE(F 1,0.609),和基于训练的合奏(F 1,0.585mmol)。在梅奥数据集,前三个是多数表决基于合奏(F 1,0.604mmol),cTAKES(F 1,0.531)和MedLEE(F 1,0.527mmol)。
结论
我们的研究表明,自然语言处理的集成可以改善单个系统的通用表型概念识别和患者特定表型概念识别。在单独的NLP系统中,当将每个单独的系统应用于最初设计的数据集中时,它们的性能最佳。但是,组合多个NLP系统以创建一个整体通常可以提高性能。具体而言,该集成可以提高跨不同队列和任务的结果可重复性,因此与单个NLP系统相比,可提供一种更便携式的表型解决方案。











































 京公网安备 11010802027423号
京公网安备 11010802027423号