当前位置:
X-MOL 学术
›
J. Mol. Recognit.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Molecular design and optimization of hepatic cancer SLP76-derived PLCγ1 SH3-binding peptide with the systematic N-substitution of peptide PXXP motif.
Journal of Molecular Recognition ( IF 2.3 ) Pub Date : 2019-08-08 , DOI: 10.1002/jmr.2806 Wenqing Tang 1 , Zhiying Zhao 2 , Chen Wang 3 , Tao Ye 3 , Biwei Yang 2
Journal of Molecular Recognition ( IF 2.3 ) Pub Date : 2019-08-08 , DOI: 10.1002/jmr.2806 Wenqing Tang 1 , Zhiying Zhao 2 , Chen Wang 3 , Tao Ye 3 , Biwei Yang 2
Affiliation
|
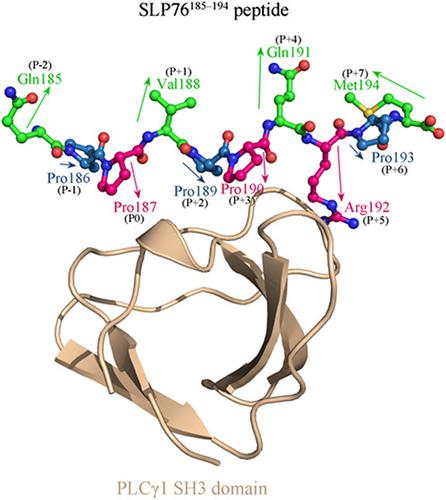
The phospholipase Cγ1 (PLCγ1) is essential for T-cell signaling and activation in hepatic cancer immune response, which has a regulatory Src homology 3 (SH3) domain that can specifically recognize and interact with the PXXP-containing decapeptide segment (185 QPPVPPQRPM194 , termed as SLP76185-194 peptide) of adaptor protein SLP76 following T-cell receptor ligation. The isolated peptide can only bind to the PLCγ1 SH3 domain with a moderate affinity due to lack of protein context support. Instead of the traditional natural residue mutagenesis that is limited by low structural diversity and shifted target specificity, we herein attempt to improve the peptide affinity by replacing the two key proline residues Pro187 and Pro190 of SLP76185-194 PXXP motif with nonnatural N-substituted amino acids, as the proline is the only endogenous N-substituted amino acid. The replacement would increase peptide flexibility but can restore peptide activity by establishing additional interactions with the domain. Structural analysis reveals that the domain pocket can be divided into a large amphipathic region and a small negatively charged region; they accommodate hydrophobic, aromatic, polar, and moderate-sized N-substituted amino acid types. A systematic replacement combination profile between the peptide residues Pro187 and Pro190 is created by structural modeling, dynamics simulation, and energetics analysis, from which six improved and two reduced N-substituted peptides as well as native SLP76185-194 peptide are identified and tested for their binding affinity to the recombinant protein of the human PLCγ1 SH3 domain using fluorescence-based assays. Two N-substituted peptides, SLP76185-194 (N-Leu187/N-Gln190) and SLP76185-194 (N-Thr187/N-Gln190), are designed to have high potency (Kd = 0.67 ± 0.18 and 1.7 ± 0.3 μM, respectively), with affinity improvement by, respectively, 8.5-fold and 3.4-fold relative to native peptide (Kd = 5.7 ± 1.2 μM).
中文翻译:

具有系统性N取代肽PXXP基序的肝癌SLP76衍生的PLCγ1SH3结合肽的分子设计和优化。
磷脂酶Cγ1(PLCγ1)对于肝癌免疫应答中的T细胞信号转导和激活至关重要,该酶具有调节性Src同源3(SH3)域,可以特异性识别含PXXP的十肽段并与之相互作用(185 QPPVPPQRPM194,称为T细胞受体连接后,衔接蛋白SLP76的SLP76185-194肽)。由于缺乏蛋白质背景支持,分离的肽只能以中等亲和力与PLCγ1SH3结构域结合。代替由低结构多样性和改变的靶标特异性所限制的传统天然残基诱变,我们在本文中尝试通过用非天然的N-取代氨基酸取代SLP76185-194 PXXP基序的两个关键脯氨酸残基Pro187和Pro190来改善肽的亲和力,因为脯氨酸是唯一的内源性N-取代氨基酸。取代将增加肽的柔性,但是可以通过与结构域建立额外的相互作用来恢复肽的活性。结构分析表明,畴袋可分为大的两亲区域和小的带负电荷的区域。它们可容纳疏水,芳香,极性和中等大小的N-取代氨基酸类型。通过结构建模,动力学模拟和能量学分析,在肽残基Pro187和Pro190之间建立了系统的置换组合概况,从中鉴定出6个改良的N和2个还原的N-取代的肽以及SLP76185-194天然肽,并对其进行了测试。使用基于荧光的分析方法对人PLCγ1SH3结构域重组蛋白的结合亲和力。
更新日期:2019-08-08
中文翻译:

具有系统性N取代肽PXXP基序的肝癌SLP76衍生的PLCγ1SH3结合肽的分子设计和优化。
磷脂酶Cγ1(PLCγ1)对于肝癌免疫应答中的T细胞信号转导和激活至关重要,该酶具有调节性Src同源3(SH3)域,可以特异性识别含PXXP的十肽段并与之相互作用(185 QPPVPPQRPM194,称为T细胞受体连接后,衔接蛋白SLP76的SLP76185-194肽)。由于缺乏蛋白质背景支持,分离的肽只能以中等亲和力与PLCγ1SH3结构域结合。代替由低结构多样性和改变的靶标特异性所限制的传统天然残基诱变,我们在本文中尝试通过用非天然的N-取代氨基酸取代SLP76185-194 PXXP基序的两个关键脯氨酸残基Pro187和Pro190来改善肽的亲和力,因为脯氨酸是唯一的内源性N-取代氨基酸。取代将增加肽的柔性,但是可以通过与结构域建立额外的相互作用来恢复肽的活性。结构分析表明,畴袋可分为大的两亲区域和小的带负电荷的区域。它们可容纳疏水,芳香,极性和中等大小的N-取代氨基酸类型。通过结构建模,动力学模拟和能量学分析,在肽残基Pro187和Pro190之间建立了系统的置换组合概况,从中鉴定出6个改良的N和2个还原的N-取代的肽以及SLP76185-194天然肽,并对其进行了测试。使用基于荧光的分析方法对人PLCγ1SH3结构域重组蛋白的结合亲和力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号