当前位置:
X-MOL 学术
›
J. Mol. Recognit.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integration of multiple computer modeling software programs for characterization of a brain natriuretic peptide sandwich DNA aptamer complex.
Journal of Molecular Recognition ( IF 2.3 ) Pub Date : 2019-08-16 , DOI: 10.1002/jmr.2809 John G Bruno 1
Journal of Molecular Recognition ( IF 2.3 ) Pub Date : 2019-08-16 , DOI: 10.1002/jmr.2809 John G Bruno 1
Affiliation
|
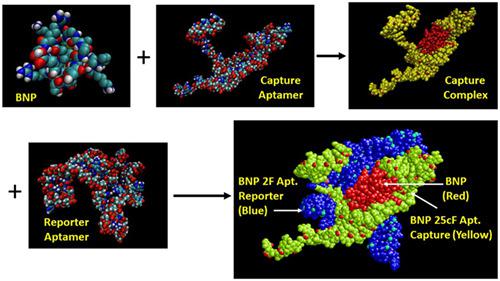
Several molecular modeling programs including Pep-Fold 3, Vienna RNA, RNA Composer, Avogadro, PatchDock, RasMol, and VMD were used to define the three-dimensional and basic binding characteristics of an extant sandwich DNA aptamer assay complex for human brain natriuretic peptide (BNP). In particular, the theoretical question of demonstrating likely binding of 72 base capture and reporter aptamers to at least two separate "epitopes" or binding sites on the small 32-amino acid BNP target was addressed, and the data support the existence of separate aptamer binding sites on BNP. The binding model was based on first docking BNP to the capture aptamer based on shape complementarity with PatchDock, followed by docking the capture aptamer-BNP complex with the reporter aptamer in PatchDock. Although, shape complementarity clearly dominated this binding model and aptamers are known to be somewhat flexible, the model demonstrates hydrogen bond stabilization within each of the two different aptamers and between the aptamers and the BNP target, thus suggesting a strong binding and high affinity sandwich assay that matches the author's former published assay results (Bruno et al., Microchem. J. 2014;115:32-38) with subpicogram per milliliter sensitivity and good specificity. Other aspects such as capture and reporter aptamer interactions in the absence of BNP are illustrated and suggest means for potentially improving the existing assay by truncating the capture and reporter aptamers where they overlap to further decrease background signal levels.
中文翻译:

集成了多个用于表征脑钠肽三明治DNA适体复合物的计算机建模软件程序。
包括Pep-Fold 3,Vienna RNA,RNA Composer,Avogadro,PatchDock,RasMol和VMD在内的几种分子建模程序用于定义现存的用于人脑利钠肽的三明治DNA适体测定复合物的三维和基本结合特征(法国巴黎银行(BNP)。特别地,解决了证明72个碱基捕获和报道适体可能结合至至少两个单独的“表位”或32个氨基酸小BNP靶标上的结合位点的理论问题,并且数据支持存在单独的适体结合BNP上的网站。结合模型基于首先基于形状互补性将BNP对接至捕获适体与PatchDock,然后将捕获适体-BNP复合物与报告适体对接在PatchDock中。虽然,形状互补性显然主导了该结合模型,并且已知适体具有一定的柔性,该模型证明了两种不同适体中的每一个以及适体和BNP靶之间的氢键稳定,从而表明了一种强结合和高亲和力的夹心测定作者先前发表的化验结果(Bruno et al。,Microchem。J.2014; 115:32-38)具有每毫升亚微微克级的灵敏度和良好的特异性。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分以进一步降低背景信号水平的潜在改进现有测定的手段。该模型证明了在两个不同的适体中的每个以及在适体和BNP靶之间的氢键稳定,因此表明了一种强结合和高亲和力的夹心测定,与作者先前发表的测定结果相符(Bruno等人,Microchem.J。 2014; 115:32-38),每毫升亚皮下灵敏度高,特异性好。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分来潜在地改善现有测定的手段,其中捕获和报告适体重叠以进一步降低背景信号水平。该模型证明了在两个不同的适体中的每个以及在适体和BNP靶之间的氢键稳定,因此表明了一种强结合和高亲和力的夹心测定,与作者先前发表的测定结果相符(Bruno等人,Microchem.J。 2014; 115:32-38),每毫升亚皮下灵敏度高,特异性好。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分以进一步降低背景信号水平的潜在改进现有测定的手段。以前发表的测定结果(Bruno et al。,Microchem。J.2014; 115:32-38),每毫升亚皮下灵敏度和良好的特异性。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分以进一步降低背景信号水平的潜在改进现有测定的手段。以前发表的测定结果(Bruno et al。,Microchem。J.2014; 115:32-38),每毫升亚皮下灵敏度和良好的特异性。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分以进一步降低背景信号水平的潜在改进现有测定的手段。
更新日期:2019-08-16
中文翻译:

集成了多个用于表征脑钠肽三明治DNA适体复合物的计算机建模软件程序。
包括Pep-Fold 3,Vienna RNA,RNA Composer,Avogadro,PatchDock,RasMol和VMD在内的几种分子建模程序用于定义现存的用于人脑利钠肽的三明治DNA适体测定复合物的三维和基本结合特征(法国巴黎银行(BNP)。特别地,解决了证明72个碱基捕获和报道适体可能结合至至少两个单独的“表位”或32个氨基酸小BNP靶标上的结合位点的理论问题,并且数据支持存在单独的适体结合BNP上的网站。结合模型基于首先基于形状互补性将BNP对接至捕获适体与PatchDock,然后将捕获适体-BNP复合物与报告适体对接在PatchDock中。虽然,形状互补性显然主导了该结合模型,并且已知适体具有一定的柔性,该模型证明了两种不同适体中的每一个以及适体和BNP靶之间的氢键稳定,从而表明了一种强结合和高亲和力的夹心测定作者先前发表的化验结果(Bruno et al。,Microchem。J.2014; 115:32-38)具有每毫升亚微微克级的灵敏度和良好的特异性。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分以进一步降低背景信号水平的潜在改进现有测定的手段。该模型证明了在两个不同的适体中的每个以及在适体和BNP靶之间的氢键稳定,因此表明了一种强结合和高亲和力的夹心测定,与作者先前发表的测定结果相符(Bruno等人,Microchem.J。 2014; 115:32-38),每毫升亚皮下灵敏度高,特异性好。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分来潜在地改善现有测定的手段,其中捕获和报告适体重叠以进一步降低背景信号水平。该模型证明了在两个不同的适体中的每个以及在适体和BNP靶之间的氢键稳定,因此表明了一种强结合和高亲和力的夹心测定,与作者先前发表的测定结果相符(Bruno等人,Microchem.J。 2014; 115:32-38),每毫升亚皮下灵敏度高,特异性好。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分以进一步降低背景信号水平的潜在改进现有测定的手段。以前发表的测定结果(Bruno et al。,Microchem。J.2014; 115:32-38),每毫升亚皮下灵敏度和良好的特异性。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分以进一步降低背景信号水平的潜在改进现有测定的手段。以前发表的测定结果(Bruno et al。,Microchem。J.2014; 115:32-38),每毫升亚皮下灵敏度和良好的特异性。示出了诸如在不存在BNP的情况下的捕获和报告适体相互作用的其他方面,并提出了通过截短捕获和报告适体重叠的部分以进一步降低背景信号水平的潜在改进现有测定的手段。











































 京公网安备 11010802027423号
京公网安备 11010802027423号