当前位置:
X-MOL 学术
›
Comp. Biochem. Physiol. D Genom. Proteom.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Transcriptome analysis and weighted gene co-expression network reveals potential genes responses to heat stress in turbot Scophthalmus maximus.
Comparative Biochemistry and Physiology D: Genomics & Proteomics ( IF 2.2 ) Pub Date : 2019-11-01 , DOI: 10.1016/j.cbd.2019.100632 Zhihui Huang 1 , Aijun Ma 1 , Shuangshuang Yang 1 , Xiaofei Liu 1 , Tingting Zhao 1 , Jinsheng Zhang 1 , Xin-An Wang 1 , Zhibin Sun 1 , Zhifeng Liu 1 , Rongjing Xu 2
Comparative Biochemistry and Physiology D: Genomics & Proteomics ( IF 2.2 ) Pub Date : 2019-11-01 , DOI: 10.1016/j.cbd.2019.100632 Zhihui Huang 1 , Aijun Ma 1 , Shuangshuang Yang 1 , Xiaofei Liu 1 , Tingting Zhao 1 , Jinsheng Zhang 1 , Xin-An Wang 1 , Zhibin Sun 1 , Zhifeng Liu 1 , Rongjing Xu 2
Affiliation
|
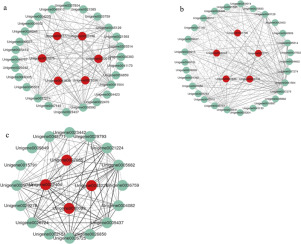
Turbot (Scophthalmus maximus) is an economically important marine fish cultured in China. In this study, we performed transcriptome gene expression profiling of kidney tissue in turbot exposed to heat stress (20, 23, 25 and 28 °C); control fish were maintained at 14 °C. We investigated gene relationships based on weighted gene co-expression network analysis (WGCNA). Accordingly, enrichment analyses of GO terms and KEGG pathways showed that several pathways (e.g., fat metabolism, cell apoptosis, immune system, and insulin signaling) may be involved in the response of turbot to heat stress. Moreover, via WGCNA, we identified 19 modules: the dark grey module was mainly enriched in pathways associated with fat metabolism and the FOXO and Jak-STAT signaling pathways. The ivory module was significantly enriched in the P53 signaling pathway. Furthermore, the key hub genes CBP, AKT3, CCND2, PIK3r2, SCOS3, mdm2, cyc-B, and p48 were enriched in the FOXO, Jak-STAT and P53 signaling pathways. This is the first study reporting co-expression patterns of a gene network after heat stress in marine fish. Our results may contribute to our understanding of the underlying molecular mechanism of thermal tolerance.
更新日期:2019-11-01











































 京公网安备 11010802027423号
京公网安备 11010802027423号