当前位置:
X-MOL 学术
›
Nat. Protoc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
MOWChIP-seq for low-input and multiplexed profiling of genome-wide histone modifications.
Nature Protocols ( IF 13.1 ) Pub Date : 2019-10-30 , DOI: 10.1038/s41596-019-0223-x Bohan Zhu 1 , Yuan-Pang Hsieh 1 , Travis W Murphy 1 , Qiang Zhang 1 , Lynette B Naler 1 , Chang Lu 1
Nature Protocols ( IF 13.1 ) Pub Date : 2019-10-30 , DOI: 10.1038/s41596-019-0223-x Bohan Zhu 1 , Yuan-Pang Hsieh 1 , Travis W Murphy 1 , Qiang Zhang 1 , Lynette B Naler 1 , Chang Lu 1
Affiliation
|
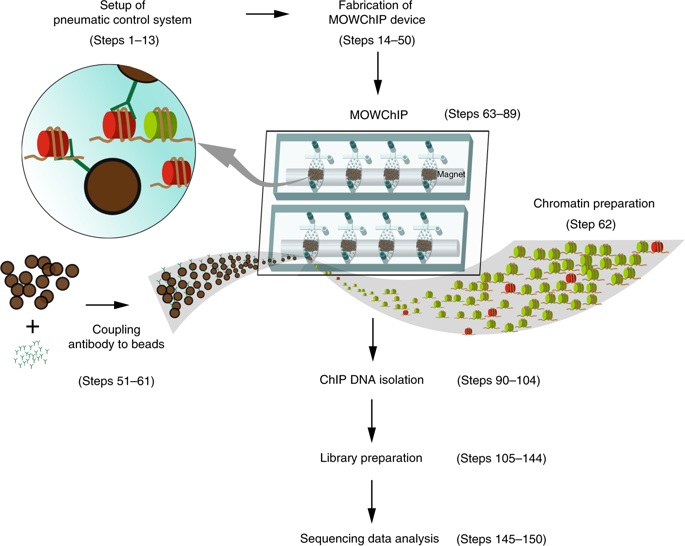
Epigenetic mechanisms such as histone modifications play critical roles in adaptive tuning of chromatin structures. Profiling of various histone modifications at the genome scale using tissues from animal and human samples is an important step for functional studies of epigenomes and epigenomics-based precision medicine. Because the profile of a histone mark is highly specific to a cell type, cell isolation from tissues is often necessary to generate a homogeneous cell population, and such operations tend to yield a low number of cells. In addition, high-throughput processing is often desirable because of the multiplexity of histone marks of interest and the large quantity of samples in a hospital setting. In this protocol, we provide detailed instructions for device fabrication, setup, and operation of microfluidic oscillatory washing-based chromatin immunoprecipitation followed by sequencing (MOWChIP-seq) for profiling of histone modifications using as few as 100 cells per assay with a throughput as high as eight assays in one run. MOWChIP-seq operation involves flowing of chromatin fragments through a packed bed of antibody-coated beads, followed by vigorous microfluidic oscillatory washing. Our process is semi-automated to reduce labor and improve reproducibility. Using one eight-unit device, it takes 2 d to produce eight sequencing libraries from chromatin samples. The technology is scalable. We used the protocol to study a number of histone modifications in various types of mouse and human tissues. The protocol can be conducted by a user who is familiar with molecular biology procedures and has basic engineering skills.
中文翻译:

MOWChIP-seq 用于全基因组组蛋白修饰的低输入和多重分析。
组蛋白修饰等表观遗传机制在染色质结构的适应性调节中发挥着关键作用。使用来自动物和人类样本的组织在基因组规模上分析各种组蛋白修饰是表观基因组功能研究和基于表观基因组学的精准医学的重要一步。因为组蛋白标记的轮廓对细胞类型具有高度特异性,从组织中分离细胞通常是产生同质细胞群所必需的,而这种操作往往会产生少量的细胞。此外,由于感兴趣的组蛋白标记的多样性和医院环境中的大量样本,通常需要高通量处理。在本协议中,我们提供了有关设备制造、设置、和操作基于微流体振荡洗涤的染色质免疫沉淀,然后进行测序(MOWChIP-seq),用于对组蛋白修饰进行分析,每次测定使用少至 100 个细胞,一次运行的通量高达 8 次测定。MOWChIP-seq 操作涉及染色质片段流过抗体包被珠的填充床,然后进行剧烈的微流体振荡洗涤。我们的流程是半自动化的,以减少劳动力并提高可重复性。使用一个八单元设备,从染色质样本中生成八个测序文库需要 2 天。该技术是可扩展的。我们使用该协议研究了各种类型的小鼠和人体组织中的一些组蛋白修饰。该协议可由熟悉分子生物学程序并具有基本工程技能的用户执行。
更新日期:2019-11-01
中文翻译:

MOWChIP-seq 用于全基因组组蛋白修饰的低输入和多重分析。
组蛋白修饰等表观遗传机制在染色质结构的适应性调节中发挥着关键作用。使用来自动物和人类样本的组织在基因组规模上分析各种组蛋白修饰是表观基因组功能研究和基于表观基因组学的精准医学的重要一步。因为组蛋白标记的轮廓对细胞类型具有高度特异性,从组织中分离细胞通常是产生同质细胞群所必需的,而这种操作往往会产生少量的细胞。此外,由于感兴趣的组蛋白标记的多样性和医院环境中的大量样本,通常需要高通量处理。在本协议中,我们提供了有关设备制造、设置、和操作基于微流体振荡洗涤的染色质免疫沉淀,然后进行测序(MOWChIP-seq),用于对组蛋白修饰进行分析,每次测定使用少至 100 个细胞,一次运行的通量高达 8 次测定。MOWChIP-seq 操作涉及染色质片段流过抗体包被珠的填充床,然后进行剧烈的微流体振荡洗涤。我们的流程是半自动化的,以减少劳动力并提高可重复性。使用一个八单元设备,从染色质样本中生成八个测序文库需要 2 天。该技术是可扩展的。我们使用该协议研究了各种类型的小鼠和人体组织中的一些组蛋白修饰。该协议可由熟悉分子生物学程序并具有基本工程技能的用户执行。











































 京公网安备 11010802027423号
京公网安备 11010802027423号